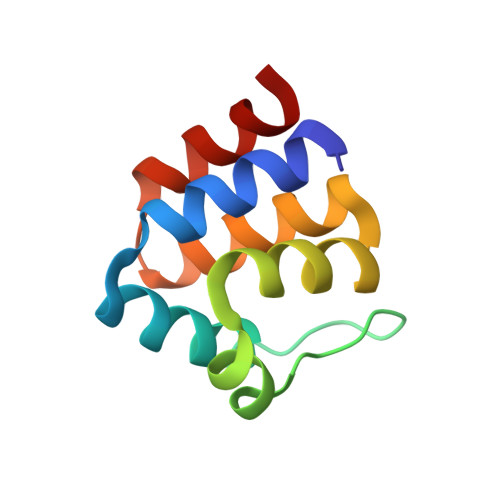
The death-domain fold of the ASC PYRIN domain, presenting a basis for PYRIN/PYRIN recognition
Liepinsh, E., Barbals, R., Dahl, E., Sharipo, A., Staub, E., Otting, G.(2003) J Mol Biol 332: 1155-1163
- PubMed: 14499617
- DOI: https://doi.org/10.1016/j.jmb.2003.07.007
- Primary Citation of Related Structures:
1UCP - PubMed Abstract:
The PYRIN domain is a conserved sequence motif identified in more than 20 human proteins with putative functions in apoptotic and inflammatory signalling pathways. The three-dimensional structure of the PYRIN domain from human ASC was determined by NMR spectroscopy. The structure determination reveals close structural similarity to death domains, death effector domains, and caspase activation and recruitment domains, although the structural alignment with these other members of the death-domain superfamily differs from previously predicted amino acid sequence alignments. Two highly positively and negatively charged surfaces in the PYRIN domain of ASC result in a strong electrostatic dipole moment that is predicted to be present also in related PYRIN domains. These results suggest that electrostatic interactions play an important role for the binding between PYRIN domains. Consequently, the previously reported binding between the PYRIN domains of ASC and ASC2/POP1 or between the zebrafish PYRIN domains of zAsc and Caspy is proposed to involve interactions between helices 2 and 3 of one PYRIN domain with helices 1 and 4 of the other PYRIN domain, in analogy to previously reported homophilic interactions between caspase activation and recruitment domains.
Organizational Affiliation:
Department of Medical Biochemistry and Biophysics, Karolinska Institute, S-17177, Stockholm, Sweden.