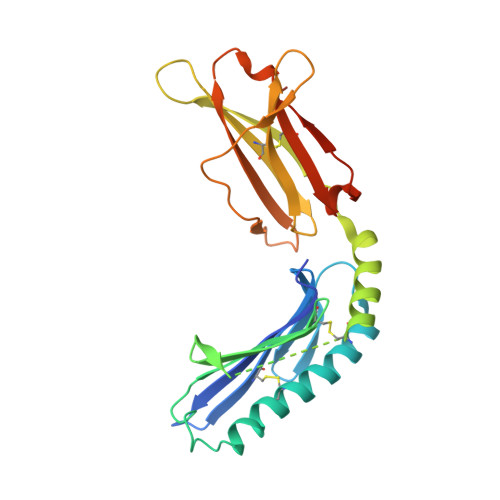
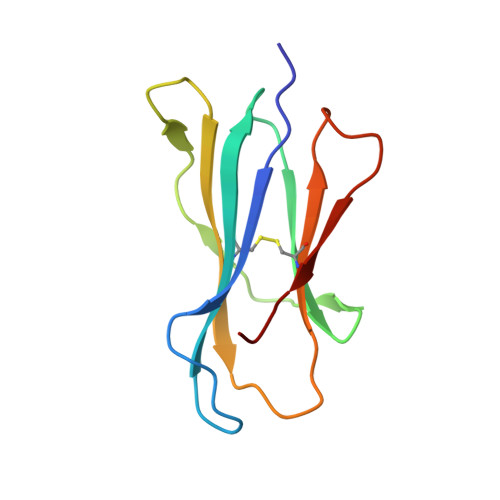
Crystal structure of the murine cytomegalovirus MHC-I homolog m144.
Natarajan, K., Hicks, A., Mans, J., Robinson, H., Guan, R., Mariuzza, R.A., Margulies, D.H.(2006) J Mol Biol 358: 157-171
- PubMed: 16500675
- DOI: https://doi.org/10.1016/j.jmb.2006.01.068
- Primary Citation of Related Structures:
1U58 - PubMed Abstract:
Large DNA viruses of the herpesvirus family produce proteins that mimic host MHC-I molecules as part of their immunoevasive strategy. The m144 glycoprotein, expressed by murine cytomegalovirus, is thought to be an MHC-I homolog whose expression prolongs viral survival in vivo by preventing natural killer cell activation. To explore the structural basis of this m144 function, we have determined the three-dimensional structure of an m144/beta2-microglobulin (beta2m) complex at 1.9A resolution. This structure reveals the canonical features of MHC-I molecules including readily identifiable alpha1, alpha2, and alpha3 domains. A unique disulfide bond links the alpha1 helix to the beta-sheet floor, explaining the known thermal stability of m144. Close juxtaposition of the alpha1 and alpha2 helices and the lack of critical residues that normally contribute to anchoring the peptide N and C termini eliminates peptide binding. A region of 13 amino acid residues, corresponding to the amino-terminal portion of the alpha2 helix, is missing in the electron density map, suggesting an area of structural flexibility that may be involved in ligand binding.
Organizational Affiliation:
Molecular Biology Section, Laboratory of Immunology, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, MD 20892-1892, USA.