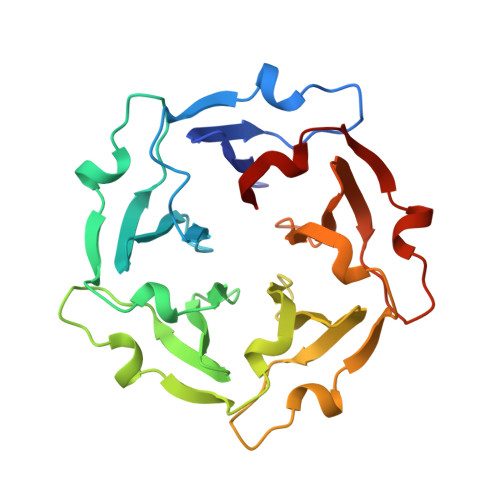
Tachylectin-2: crystal structure of a specific GlcNAc/GalNAc-binding lectin involved in the innate immunity host defense of the Japanese horseshoe crab Tachypleus tridentatus.
Beisel, H.G., Kawabata, S., Iwanaga, S., Huber, R., Bode, W.(1999) EMBO J 18: 2313-2322
- PubMed: 10228146
- DOI: https://doi.org/10.1093/emboj/18.9.2313
- Primary Citation of Related Structures:
1TL2 - PubMed Abstract:
Tachylectin-2, isolated from large granules of the hemocytes of the Japanese horseshoe crab (Tachypleus tridentatus), is a 236 amino acid protein belonging to the lectins. It binds specifically to N-acetylglucosamine and N-acetylgalactosamine and is a part of the innate immunity host defense system of the horseshoe crab. The X-ray structure of tachylectin-2 was solved at 2.0 A resolution by the multiple isomorphous replacement method and this molecular model was employed to solve the X-ray structure of the complex with N-acetylglucosamine. Tachylectin-2 is the first protein displaying a five-bladed beta-propeller structure. Five four-stranded antiparallel beta-sheets of W-like topology are arranged around a central water-filled tunnel, with the water molecules arranged as a pentagonal dodecahedron. Tachylectin-2 exhibits five virtually identical binding sites, one in each beta-sheet. The binding sites are located between adjacent beta-sheets and are made by a large loop between the outermost strands of the beta-sheets and the connecting segment from the previous beta-sheet. The high number of five binding sites within the single polypeptide chain strongly suggests the recognition of carbohydrate surface structures of pathogens with a fairly high ligand density. Thus, tachylectin-2 employs strict specificity for certain N-acetyl sugars as well as the surface ligand density for self/non-self recognition.
Organizational Affiliation:
Max-Planck-Institut für Biochemie, Am Klopferspitz 18a, 82152 Martinsried, Germany. beisel@biochem.mpg.de