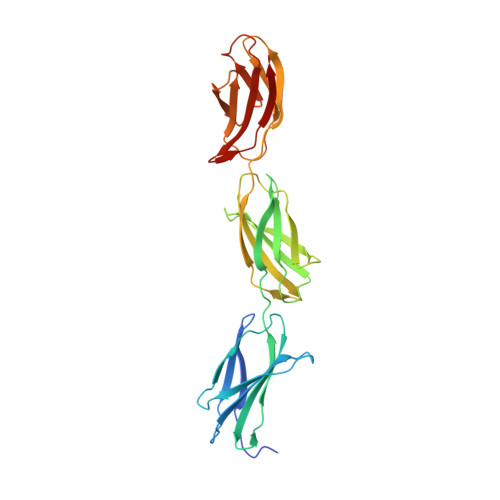
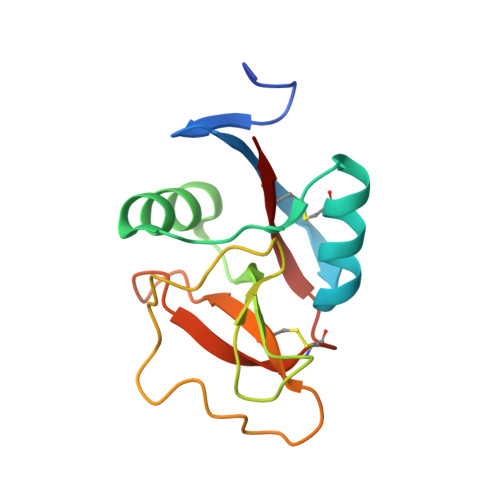
Structural basis for interactions between tenascins and lectican C-type lectin domains: evidence for a crosslinking role for tenascins
Lundell, A., Olin, A.I., Moergelin, M., al-Karadaghi, S., Aspberg, A., Logan, D.T.(2004) Structure 12: 1495-1506
- PubMed: 15296743
- DOI: https://doi.org/10.1016/j.str.2004.05.021
- Primary Citation of Related Structures:
1TDQ - PubMed Abstract:
The C-terminal G3 domains of lecticans mediate crosslinking to diverse extracellular matrix (ECM) proteins during ECM assembly, through their C-type lectin (CLD) subdomains. The structure of the rat aggrecan CLD in a Ca(2+)-dependent complex with fibronectin type III repeats 3-5 of rat tenascin-R provides detailed support for such crosslinking. The CLD loops bind Ca2+ like other CLDs, but no carbohydrate binding is observed or possible. This is thus the first example of a direct Ca(2+)-dependent protein-protein interaction of a CLD. Surprisingly, tenascin-R does not coordinate the Ca2+ ions directly. Electron microscopy confirms that full-length tenascin-R and tenascin-C crosslink hyaluronan-aggrecan complexes. The results are significant for the binding of all lectican CLDs to tenascin-R and tenascin-C. Comparison of the protein interaction surface with that of P-selectin in complex with the PGSL-1 peptide suggests that direct protein-protein interactions of Ca(2+)-binding CLDs may be more widespread than previously appreciated.
Organizational Affiliation:
Department of Molecular Biophysics, Lund University, Box 124, S-221 00 Lund, Sweden.