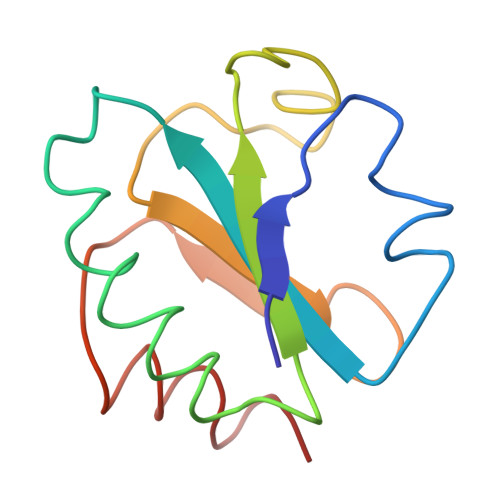
Three-dimensional structure of Escherichia coli thioredoxin-S2 to 2.8 A resolution.
Holmgren, A., Soderberg, B.O., Eklund, H., Branden, C.I.(1975) Proc Natl Acad Sci U S A 72: 2305-2309
- PubMed: 1094461
- DOI: https://doi.org/10.1073/pnas.72.6.2305
- Primary Citation of Related Structures:
1SRX - PubMed Abstract:
The three-dimensional structure of the electron transport protein thioredoxin-S2 from E. coli has been determined from a 2.8 A resolution electron density map. The molecule is built up of a central core of three parallel and two antiparallel strands of pleated sheet surrounded by four helices. Thr residues involved in the active center 14-membered disulfide ring of thioredoxin form a protrusion between one of the helices and the middle strand of the pleated sheet. This region of the molecule, comprising two parallel strands joined by the protrusion and a helix, is structurally very similar to corresponding functionally important regions in the nucleotide-binding domains of flavodoxin and the dehydrogenases. The molecule has about 75% of the residues in well-defined secondary structures. The structure indicates that the carboxy-terminal third of the molecule forms an independent folding unit consisting of two strands of antiparallel pleated sheet and a terminal alpha-helix. This agress with the noncovalent reconstitution experiments from thioredoxin peptide fragments. Thioredoxin is an example of a protein with the active center located on a protrusion rather than in a cleft, thus demonstrating the existence of male proteins.