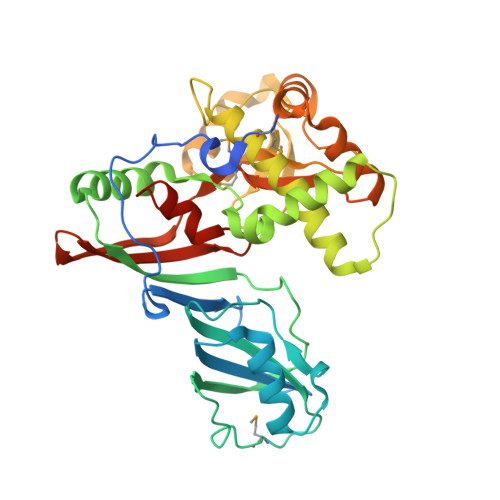
Crystal structure of the highly divergent pseudouridine synthase TruD reveals a circular permutation of a conserved fold.
Hoang, C., Ferre-D'Amare, A.R.(2004) RNA 10: 1026-1033
- PubMed: 15208439
- DOI: https://doi.org/10.1261/rna.7240504
- Primary Citation of Related Structures:
1SB7 - PubMed Abstract:
The pseudouridine (Psi) synthases Pus7p and TruD define a family of RNA-modifying enzymes with no sequence similarity to previously characterized Psi synthases. The 2.2 A resolution structure of Escherichia coli TruD reveals a U-shaped molecule with a catalytic domain that superimposes closely on that of other Psi synthases. A domain that appears to be unique to TruD/Pus7p family enzymes hinges over the catalytic domain, possibly serving to clasp the substrate RNAs. The active site comprises residues that are conserved in other Psi synthases, although at least one comes from a structurally distinct part of the protein. Remarkably, the connectivity of the structural elements of the TruD catalytic domain is a circular permutation of that of its paralogs. Because the sequence of the permuted segment, a beta-strand that bisects the catalytic domain, is conserved among orthologs from bacteria, archaea and eukarya, the permutation likely happened early in evolution.
Organizational Affiliation:
Division of Basic Sciences, Fred Hutchinson Cancer Research Center, 1100 Fairview Avenue North, Seattle, WA 98109-1024, USA.