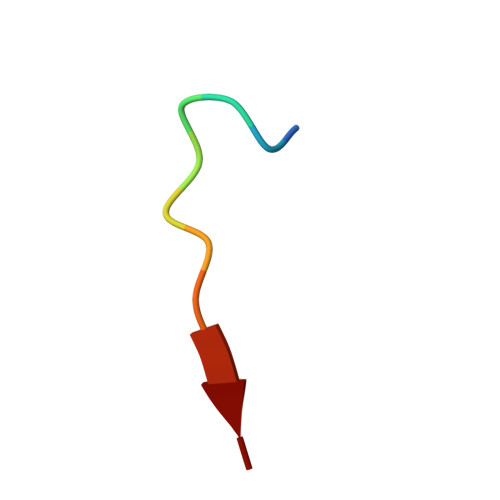
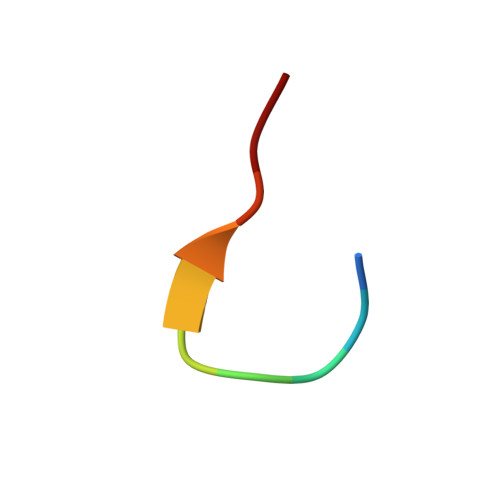
Structure of thermolysin cleaved microcin J25: extreme stability of a two-chain antimicrobial peptide devoid of covalent links
Rosengren, K.J., Blond, A., Afonso, C., Tabet, J.C., Rebuffat, S., Craik, D.J.(2004) Biochemistry 43: 4696-4702
- PubMed: 15096038
- DOI: https://doi.org/10.1021/bi0361261
- Primary Citation of Related Structures:
1S7P - PubMed Abstract:
The structure of a two-chain peptide formed by the treatment of the potent antimicrobial peptide microcin J25 (MccJ25) with thermolysin has been characterized by NMR spectroscopy and mass spectrometry. The native peptide is 21 amino acids in size and has the remarkable structural feature of a ring formed by linkage of the side chain of Glu8 to the N-terminus that is threaded by the C-terminal tail of the peptide. Thermolysin cleaves the peptide at the Phe10-Val11 amide bond, but the threading of the C-terminus through the N-terminal ring is so tight that the resultant two chains remain associated both in the solution and in the gas phases. The three-dimensional structure of the thermolysin-cleaved peptide derived using NMR spectroscopy and simulated annealing calculations has a well-defined core that comprises the N-terminal ring and the threading C-terminal tail. In contrast to the well-defined core, the newly formed termini at residues Phe10 and Val11 are disordered in solution. The C-terminal tail is associated to the ring both by hydrogen bonds stabilizing a short beta-sheet and by hydrophobic interactions. Moreover, unthreading of the tail through the ring is prevented by the bulky side chains of Phe19 and Tyr20, which flank the octapeptide ring. This noncovalent two-peptide complex that has a remarkable stability in solution and in highly denaturing conditions and that survives in the gas phase is the first example of such a two-chain peptide lacking disulfide or interchain covalent bonds.
Organizational Affiliation:
Institute for Molecular Bioscience, University of Queensland, Brisbane, QLD 4072, Australia.