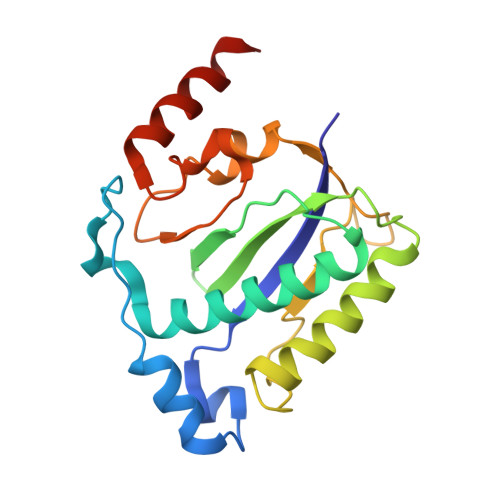
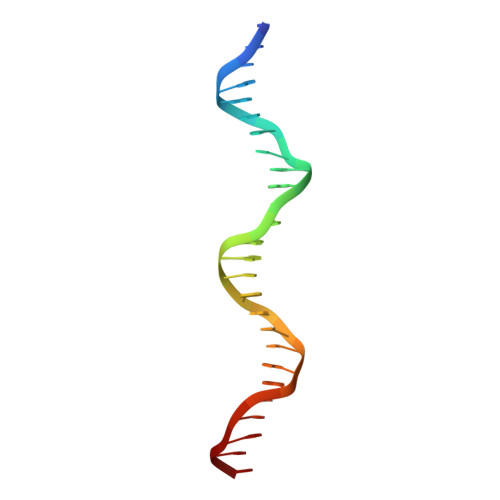
The nuclease domain of adeno-associated virus rep coordinates replication initiation using two distinct DNA recognition interfaces.
Hickman, A.B., Ronning, D.R., Perez, Z.N., Kotin, R.M., Dyda, F.(2004) Mol Cell 13: 403-414
- PubMed: 14967147
- DOI: https://doi.org/10.1016/s1097-2765(04)00023-1
- Primary Citation of Related Structures:
1RZ9, 1UUT - PubMed Abstract:
Integration into a particular location in human chromosomes is a unique property of the adeno-associated virus (AAV). This reaction requires the viral Rep protein and AAV origin sequences. To understand how Rep recognizes DNA, we have determined the structures of the Rep endonuclease domain separately complexed with two DNA substrates: the Rep binding site within the viral inverted terminal repeat and one of the terminal hairpin arms. At the Rep binding site, five Rep monomers bind five tetranucleotide direct repeats; each repeat is recognized by two Rep monomers from opposing faces of the DNA. Stem-loop binding involves a protein interface on the opposite side of the molecule from the active site where ssDNA is cleaved. Rep therefore has three distinct binding sites within its endonuclease domain for its different DNA substrates. Use of these different interfaces generates the structural asymmetry necessary to regulate later events in viral replication and integration.
Organizational Affiliation:
Laboratory of Molecular Biology, National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health, Bethesda, MD 20892 USA.