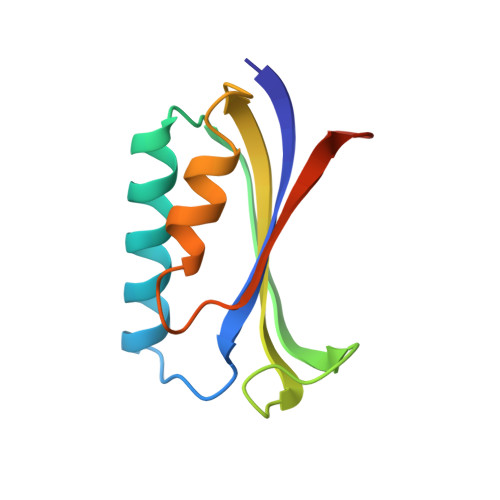
Crystal structure of the ribosomal protein S6 from Thermus thermophilus.
Lindahl, M., Svensson, L.A., Liljas, A., Sedelnikova, S.E., Eliseikina, I.A., Fomenkova, N.P., Nevskaya, N., Nikonov, S.V., Garber, M.B., Muranova, T.A., Rykonova, A.I., Amons, R.(1994) EMBO J 13: 1249-1254
- PubMed: 8137808
- DOI: https://doi.org/10.2210/pdb1ris/pdb
- Primary Citation of Related Structures:
1RIS - PubMed Abstract:
The amino acid sequence and crystal structure of the ribosomal protein S6 from the small ribosomal subunit of Thermus thermophilus have been determined. S6 is a small protein with 101 amino acid residues. The 3D structure, which was determined to 2.0 A resolution, consists of a four-stranded anti-parallel beta-sheet with two alpha-helices packed on one side. Similar folding patterns have been observed for other ribosomal proteins and may suggest an original RNA-interacting motif. Related topologies are also found in several other nucleic acid-interacting proteins and based on the assumption that the structure of the ribosome was established early in the molecular evolution, the possibility that an ancestral RNA-interacting motif in ribosomal proteins is the evolutionary origin for the nucleic acid-interacting domain in large classes of ribonucleic acid binding proteins should be considered.
Organizational Affiliation:
Chemical Center, University of Lund, Sweden.