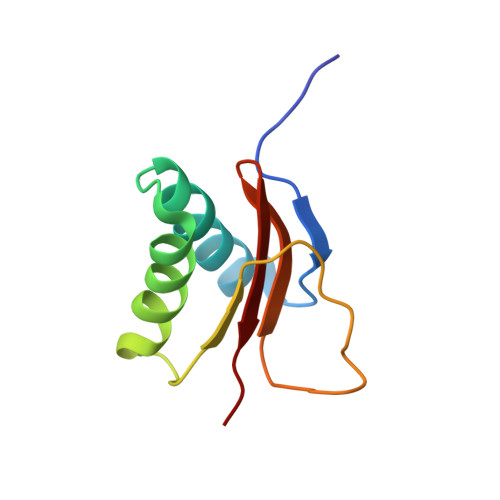
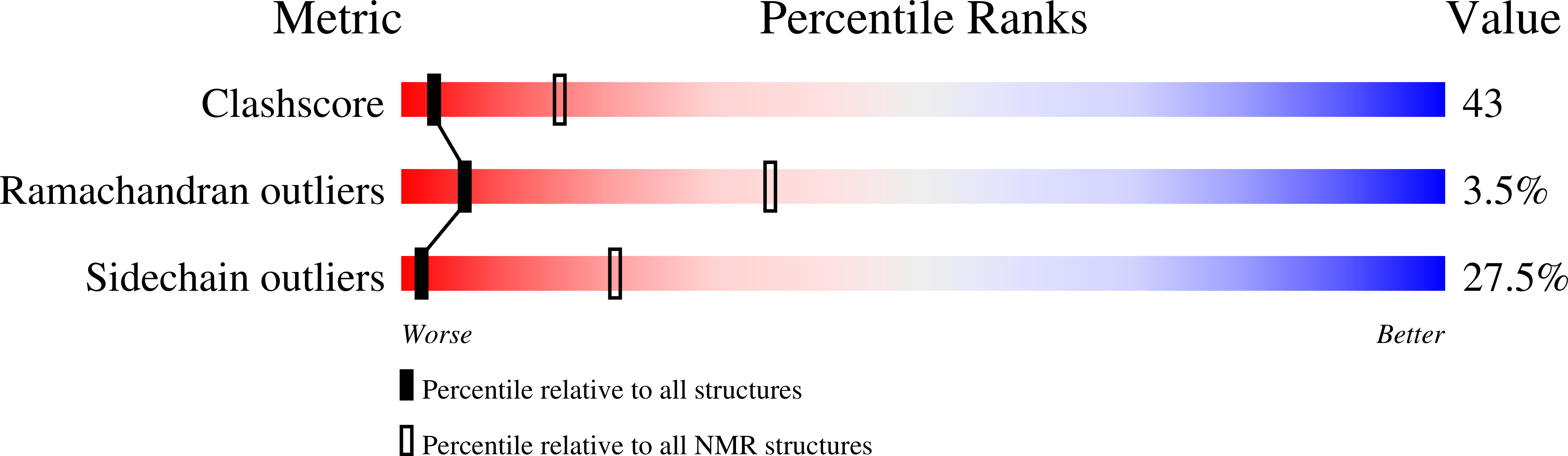The solution structure of the pH-induced monomer of dynein light-chain LC8 from Drosophila.
Makokha, M., Huang, Y.J., Montelione, G., Edison, A.S., Barbar, E.(2004) Protein Sci 13: 727-734
- PubMed: 14767079
- DOI: https://doi.org/10.1110/ps.03462204
- Primary Citation of Related Structures:
1RHW - PubMed Abstract:
The structure of Drosophila LC8 pH-induced monomer has been determined by NMR spectroscopy using the program AutoStructure. The structure at pH 3 and 30 degrees C is similar to the individual subunits of mammalian LC8 dimer with the exception that a beta strand, which crosses between monomers to form an intersubunit beta-sheet in the dimer, is a flexible loop with turnlike conformations in the monomer. Increased flexibility in the interface region relative to the rest of the protein is confirmed by dynamic measurements based on (15)N relaxation. Comparison of the monomer and dimer structures indicates that LC8 is not a domain swapped dimer.
Organizational Affiliation:
Department of Biochemistry and Biophysics, Ohio University, Athens, 45701-2979, USA.