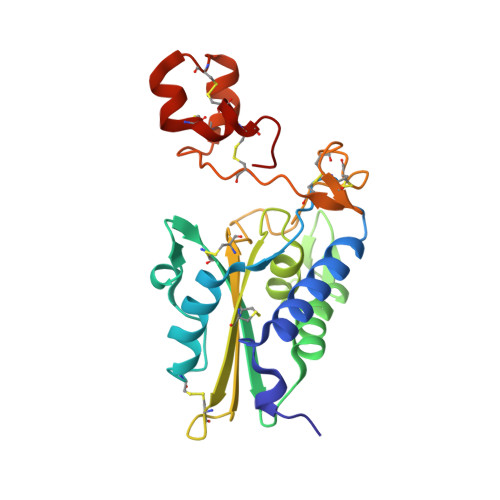
Crystal structure of cysteine-rich secretory protein stecrisp reveals the cysteine-rich domain has a K+-channel inhibitor-like fold.
Guo, M., Teng, M., Niu, L., Liu, Q., Huang, Q., Hao, Q.(2004) J Biol Chem 280: 12405-12412
- PubMed: 15596436
- DOI: https://doi.org/10.1074/jbc.M413566200
- Primary Citation of Related Structures:
1RC9 - PubMed Abstract:
Stecrisp from Trimeresurus stejnegeri snake venom belongs to a family of cysteine-rich secretory proteins (CRISP) that have various functions related to sperm-egg fusion, innate host defense, and the blockage of ion channels. Here we present the crystal structure of stecrisp refined to 1.6-angstrom resolution. It shows that stecrisp contains three regions, namely a PR-1 (pathogenesis-related proteins of group1) domain, a hinge, and a cysteine-rich domain (CRD). A conformation of solvent-exposed and -conserved residues (His60, Glu75, Glu96, and His115) in the PR-1 domain similar to that of their counterparts in homologous structures suggests they may share some molecular mechanism. Three flexible loops of hypervariable sequence surrounding the possible substrate binding site in the PR-1 domain show an evident difference in homologous structures, implying that a great diversity of species- and substrate-specific interactions may be involved in recognition and catalysis. The hinge is fixed by two crossed disulfide bonds formed by four of ten characteristic cysteines in the carboxyl-terminal region and is important for stabilizing the N-terminal PR-1 domain. Spatially separated from the PR-1 domain, CRD possesses a similar fold with two K+ channel inhibitors (Bgk and Shk). Several candidates for the possible functional sites of ion channel blocking are located in a solvent-exposed loop in the CRD. The structure of stecrisp will provide a prototypic architecture for a structural and functional exploration of the diverse members of the CRISP family.
Organizational Affiliation:
Key Laboratory of Structural Biology, Department of Molecular and Cell Biology, School of Life Sciences, Hefei National Laboratory for Physical Sciences at Microscale, University of Science and Technology of China, China.