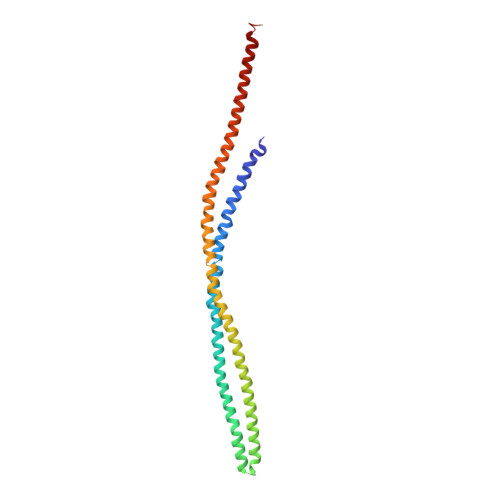Four-helical-bundle structure of the cytoplasmic domain of a serine chemotaxis receptor.
Kim, K.K., Yokota, H., Kim, S.H.(1999) Nature 400: 787-792
- PubMed: 10466731
- DOI: https://doi.org/10.1038/23512
- Primary Citation of Related Structures:
1QU7 - PubMed Abstract:
The bacterial chemotaxis receptors are transmembrane receptors with a simple signalling pathway which has elements relevant to the general understanding of signal recognition and transduction across membranes, how signals are relayed between molecules in a pathway, and how adaptation to a persistent signal is achieved. In contrast to many mammalian receptors which signal by oligomerizing upon ligand binding, the chemotaxis receptors are dimeric even in the absence of their ligands, and their signalling does not depend on a monomer-dimer equilibrium. Bacterial chemotaxis receptors are composed of a ligand-binding domain, a transmembrane domain consisting of two helices TM1 and TM2, and a cytoplasmic domain. All known bacterial chemotaxis receptors have a highly conserved cytoplasmic domain, which unites signals from different ligand domains into a single signalling pathway to flagella motors. Here we report the crystal structure of the cytoplasmic domain of a serine chemotaxis receptor of Escherichia coli, which reveals a 200 A-long coiled-coil of two antiparallel helices connected by a 'U-turn'. Two of these domains form a long, supercoiled, four-helical bundle in the cytoplasmic portion of the receptor.
Organizational Affiliation:
Department of Chemistry, University of California, Berkeley 94720-5230, USA.