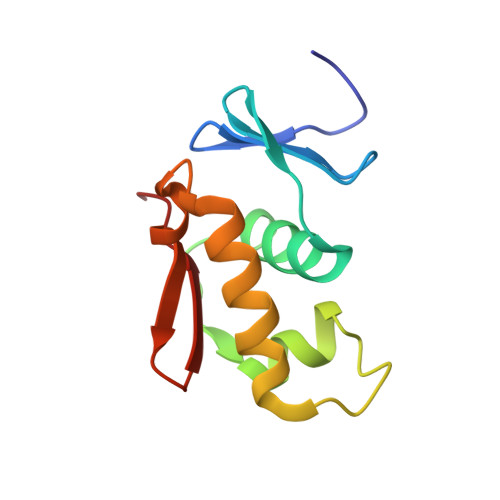
Structural comparison of the PhoB and OmpR DNA-binding/transactivation domains and the arrangement of PhoB molecules on the phosphate box.
Okamura, H., Hanaoka, S., Nagadoi, A., Makino, K., Nishimura, Y.(2000) J Mol Biol 295: 1225-1236
- PubMed: 10653699
- DOI: https://doi.org/10.1006/jmbi.1999.3379
- Primary Citation of Related Structures:
1QQI - PubMed Abstract:
PhoB is a transcriptional activator that binds to the phosphate box in the promoters of the phosphate genes of Escherichia coli. PhoB contains two functional domains, an N-terminal phosphorylation domain and a C-terminal DNA-binding/transactivation domain. Here, the three-dimensional structure of the DNA-binding/transactivation domain has been determined by NMR. It consists of an N-terminal four-stranded beta-sheet, a central three helical bundle and a C-terminal beta-hairpin. The second and third helices form a helix-turn-helix (HTH) variant containing a longer turn than the corresponding turn of the classical HTH motif. The overall architecture is very close to that of the OmpR DNA-binding/transactivation domain, however, the conformation of the long turn region of PhoB, a putative interaction site for the RNA polymerase sigma subunit, is entirely different from that of the corresponding turn of OmpR, which interacts with the alpha subunit. In addition, the third helix of PhoB is three amino acid residues longer than the corresponding helix of OmpR. The binding site of PhoB is a TGTCA sequence and the phospahte box contains the two binding sites. NMR studies of the complexes of the PhoB DNA-binding/transactivation domain bound to several different DNA molecules have revealed that two PhoB molecules bind in a tandem array on the phosphate box. In each complex of PhoB the third helix of the DNA-binding/transactivation domain is likely to recognize the TGTCA sequence from the major groove of DNA and the C-terminal beta-hairpin contacts on the minor groove of the 3' site out of the TGTCA sequence in a non-specific manner. The long turn region facing outward is likely to interact with the sigma subunit.
Organizational Affiliation:
Graduate School of Integrated Science, Yokohama City University, 22-2 Seto, Yokohama, Kanazawa-ku, 236-0027, Japan.