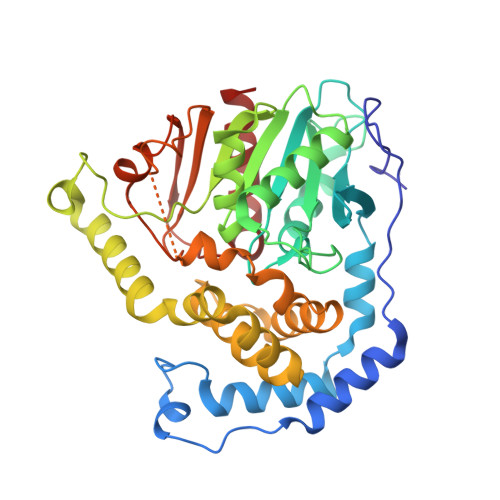
Structure of Aspergillus Niger Epoxide Hydrolase at 1.8A Resolution: Implications for the Structure and Function of the Mammalian Microsomal Class of Epoxide Hydrolases
Zou, J.-Y., Hallberg, B.M., Bergfors, T., Oesch, F., Arand, M., Mowbray, S.L., Jones, T.A.(2000) Structure 8: 111
- PubMed: 10673439
- DOI: https://doi.org/10.1016/s0969-2126(00)00087-3
- Primary Citation of Related Structures:
1QO7 - PubMed Abstract:
Epoxide hydrolases have important roles in the defense of cells against potentially harmful epoxides. Conversion of epoxides into less toxic and more easily excreted diols is a universally successful strategy. A number of microorganisms employ the same chemistry to process epoxides for use as carbon sources.
Organizational Affiliation:
Department of Cell and Molecular Biology, BMC, Uppsala University, Box 596, Uppsala, S-751 24, Sweden.