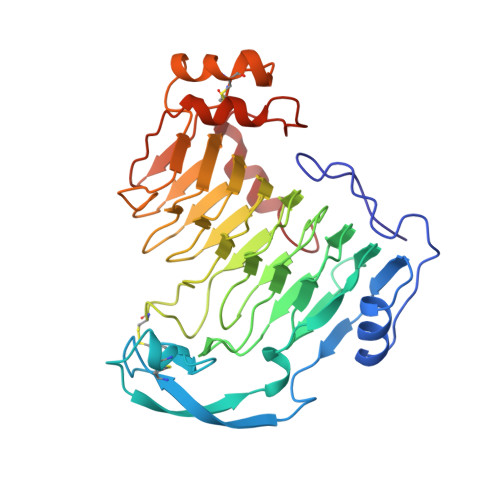
The tree-dimensional structure of aspergillus niger pectin lyase B at 1.7-A resolution.
Vitali, J., Schick, B., Kester, H.C., Visser, J., Jurnak, F.(1998) Plant Physiol 116: 69-80
- PubMed: 9449837
- DOI: https://doi.org/10.1104/pp.116.1.69
- Primary Citation of Related Structures:
1QCX - PubMed Abstract:
The three-dimensional structure of Aspergillus niger pectin lyase B (PLB) has been determined by crystallographic techniques at a resolution of 1.7 A. The model, with all 359 amino acids and 339 water molecules, refines to a final crystallographic R factor of 16.5%. The polypeptide backbone folds into a large right-handed cylinder, termed a parallel beta helix. Loops of various sizes and conformations protrude from the central helix and probably confer function. The largest loop of 53 residues folds into a small domain consisting of three antiparallel beta strands, one turn of an alpha helix, and one turn of a 3(10) helix. By comparison with the structure of Erwinia chrysanthemi pectate lyase C (PelC), the primary sequence alignment between the pectate and pectin lyase subfamilies has been corrected and the active site region for the pectin lyases deduced. The substrate-binding site in PLB is considerably less hydrophilic than the comparable PelC region and consists of an extensive network of highly conserved Trp and His residues. The PLB structure provides an atomic explanation for the lack of a catalytic requirement for Ca2+ in the pectin lyase family, in contrast to that found in the pectate lyase enzymes. Surprisingly, however, the PLB site analogous to the Ca2+ site in PelC is filled with a positive charge provided by a conserved Arg in the pectin lyases. The significance of the finding with regard to the enzymatic mechanism is discussed.
Organizational Affiliation:
Department of Biochemistry, University of California, Irvine 92512, USA.