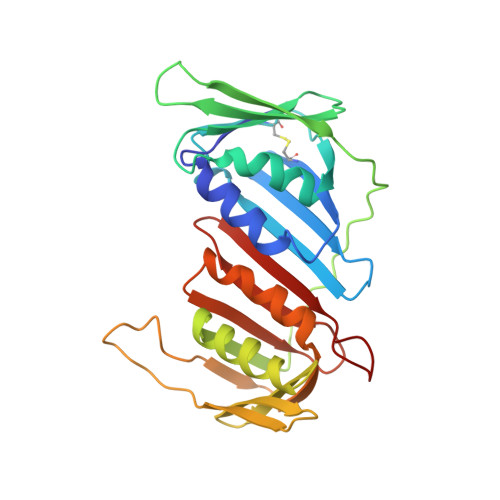
Crystal structure of the eukaryotic DNA polymerase processivity factor PCNA.
Krishna, T.S., Kong, X.P., Gary, S., Burgers, P.M., Kuriyan, J.(1994) Cell 79: 1233-1243
- PubMed: 8001157
- DOI: https://doi.org/10.1016/0092-8674(94)90014-0
- Primary Citation of Related Structures:
1PLQ, 1PLR - PubMed Abstract:
The crystal structure of the processivity factor required by eukaryotic DNA polymerase delta, proliferating cell nuclear antigen (PCNA) from S. cerevisiae, has been determined at 2.3 A resolution. Three PCNA molecules, each containing two topologically identical domains, are tightly associated to form a closed ring. The dimensions and electrostatic properties of the ring suggest that PCNA encircles duplex DNA, providing a DNA-bound platform for the attachment of the polymerase. The trimeric PCNA ring is strikingly similar to the dimeric ring formed by the beta subunit (processivity factor) of E. coli DNA polymerase III holoenzyme, with which it shares no significant sequence identity. This structural correspondence further substantiates the mechanistic connection between eukaryotic and prokaryotic DNA replication that has been suggested on biochemical grounds.
Organizational Affiliation:
Laboratories of Molecular BIophysics, Rockefeller University, New York, New York 10021.