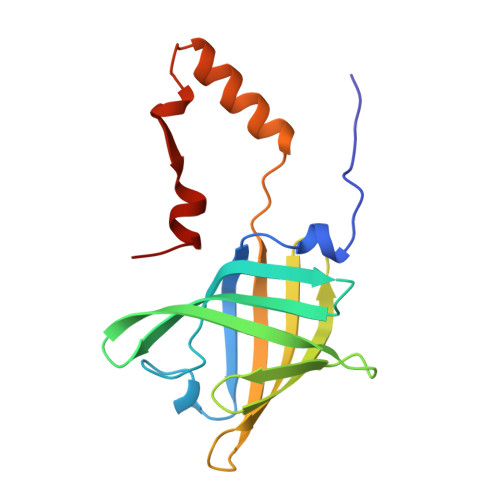
The three-dimensional structure of bovine odorant binding protein and its mechanism of odor recognition.
Bianchet, M.A., Bains, G., Pelosi, P., Pevsner, J., Snyder, S.H., Monaco, H.L., Amzel, L.M.(1996) Nat Struct Biol 3: 934-939
- PubMed: 8901871
- DOI: https://doi.org/10.1038/nsb1196-934
- Primary Citation of Related Structures:
1PBO - PubMed Abstract:
Odorant binding protein (OBP) is the major odorant binding component of mammalian nasal mucosa. The two structures of bovine OBP reported in this paper (one crystallized as purified and one soaked in the presence of a selenium-containing odorant) show that: (i) the OBP dimer is composed of two compact domains related by an approximate two-fold axis of symmetry; (ii) between residues 122 and 123 the polypeptide chains cross from one domain to the other such that each domain is formed by residues from both monomers; (iii) purified OBP already contains two bound odorant molecules (one per monomer)-odorant binding occurs by replacement of these molecules with the added odorant; and (iv) the structure of the odorant binding site can explain OBP's extraordinarily broad odorant specificity.
Organizational Affiliation:
Department of Biophysics and Biophysical Chemistry, University of Pisa, Italy. mario@neruda.med.jhu.edu