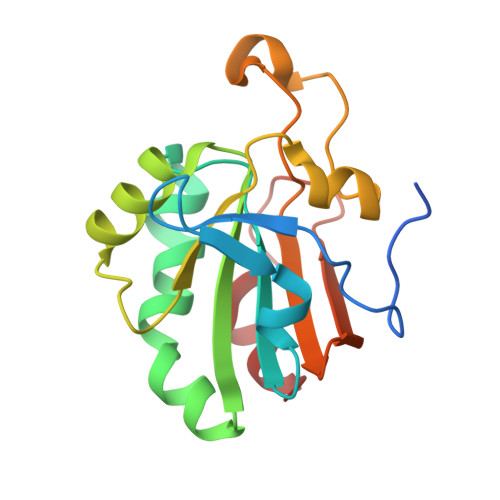
Crystal Structure of a Dimeric Oxidized Form of Human Peroxiredoxin 5
Evrard, C., Capron, A., Marchand, C., Clippe, A., Wattiez, R., Soumillion, P., Knoops, B., Declercq, J.-P.(2004) J Mol Biol 337: 1079
- PubMed: 15046979
- DOI: https://doi.org/10.1016/j.jmb.2004.02.017
- Primary Citation of Related Structures:
1OC3 - PubMed Abstract:
Peroxiredoxin 5 is the last discovered mammalian member of an ubiquitous family of peroxidases widely distributed among prokaryotes and eukaryotes. Mammalian peroxiredoxin 5 has been recently classified as an atypical 2-Cys peroxiredoxin due to the presence of a conserved peroxidatic N-terminal cysteine (Cys47) and an unconserved resolving C-terminal cysteine residue (Cys151) forming an intramolecular disulfide intermediate in the oxidized enzyme. We have recently reported the crystal structure of human peroxiredoxin 5 in its reduced form. Here, a new crystal form of human peroxiredoxin 5 is described at 2.0 A resolution. The asymmetric unit contains three polypeptide chains. Surprisingly, beside two reduced chains, the third one is oxidized although the enzyme was crystallized under initial reducing conditions in the presence of 1 mM 1,4-dithio-dl-threitol. The oxidized polypeptide chain forms an homodimer with a symmetry-related one through intermolecular disulfide bonds between Cys47 and Cys151. The formation of these disulfide bonds is accompanied by the partial unwinding of the N-terminal parts of the alpha2 helix, which, in the reduced form, contains the peroxidatic Cys47 and the alpha6 helix, which is sequentially close to the resolving residue Cys151. In each monomer of the oxidized chain, the C-terminal part including the alpha6 helix is completely reorganized and is isolated from the rest of the protein on an extended arm. In the oxidized dimer, the arm belonging to the first monomer now appears at the surface of the second subunit and vice versa.
Organizational Affiliation:
Unit of Structural Chemistry (CSTR), Université catholique de Louvain, 1 place Louis Pasteur, B-1348 Louvain-la-Neuve, Belgium.