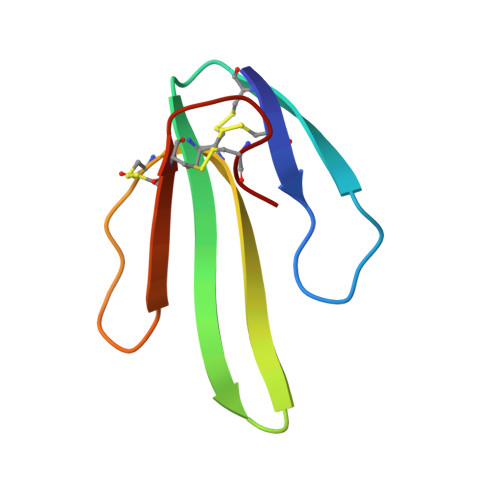
Secondary structure determination for alpha-neurotoxin from Dendroaspis polylepis polylepis based on sequence-specific 1H-nuclear-magnetic-resonance assignments.
Labhardt, A.M., Hunziker-Kwik, E.H., Wuthrich, K.(1988) Eur J Biochem 177: 295-305
- PubMed: 2847926
- DOI: https://doi.org/10.1111/j.1432-1033.1988.tb14376.x
- Primary Citation of Related Structures:
1NTX - PubMed Abstract:
Sequence-specific assignments are presented for the polypeptide backbone protons and a majority of the amino-acid-side-chain protons of alpha-neurotoxin from Dendroaspis polylepis polylepis, and individual amide proton-exchange rates with the solvent are reported. The secondary structure and the hydrogen-bonding patterns in the regular secondary structure elements are deduced from nuclear Overhauser effects and the sequence locations of the slowly exchanging amide protons. The molecule includes a three-stranded antiparallel beta-sheet, and there are indications that two additional short chain segments are arranged in an antiparallel beta-sheet. These structural elements are similar, but not identical, to either the secondary structure reported for erabutoxin b in single crystals, or the solution structure of cytotoxin CTXIIb from Naja mossambica mossambica.
Organizational Affiliation:
Institut für Molekularbiologie und Biophysik, Eidgenössische Technische Hochschule-Hönggerberg, Zürich, Switzerland.