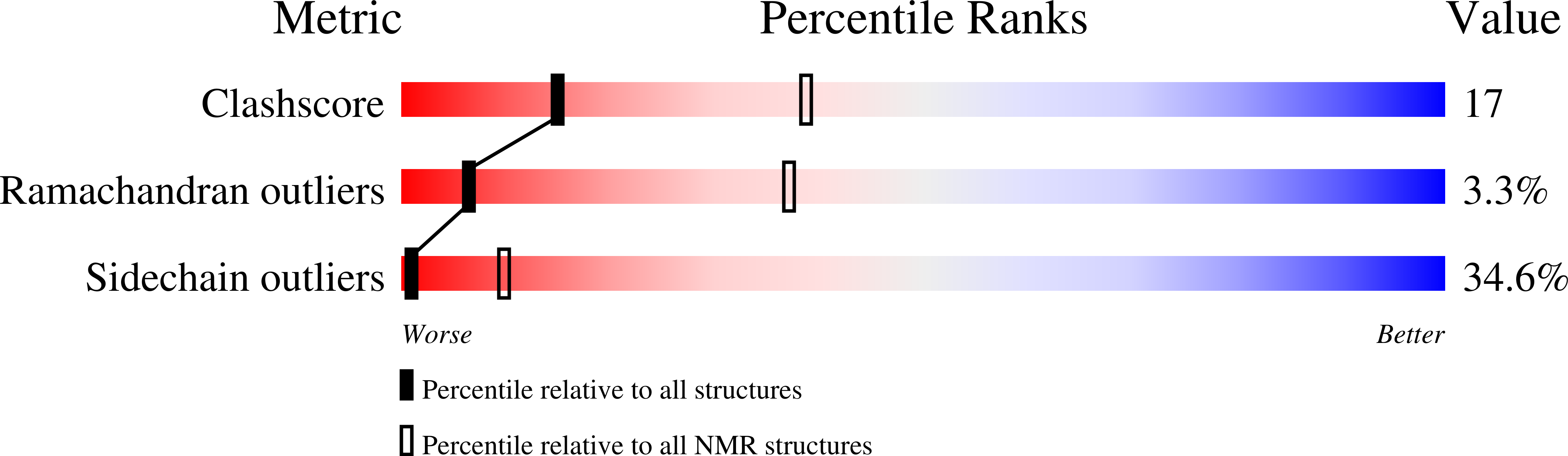A novel RNA-binding motif in influenza A virus non-structural protein 1.
Chien, C.Y., Tejero, R., Huang, Y., Zimmerman, D.E., Rios, C.B., Krug, R.M., Montelione, G.T.(1997) Nat Struct Biol 4: 891-895
- PubMed: 9360601
- DOI: https://doi.org/10.1038/nsb1197-891
- Primary Citation of Related Structures:
1NS1 - PubMed Abstract:
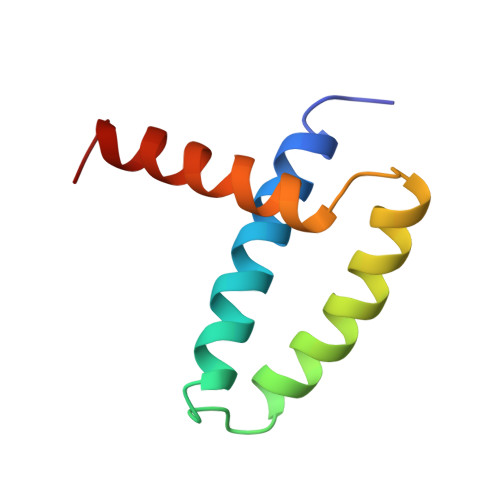
The solution NMR structure of the RNA-binding domain from influenza virus non-structural protein 1 exhibits a novel dimeric six-helical protein fold. Distributions of basic residues and conserved salt bridges of dimeric NS1(1-73) suggest that the face containing antiparallel helices 2 and 2' forms a novel arginine-rich nucleic acid binding motif.