Two-dimensional 1H-NMR study of the spatial structure of neurotoxin II from Naja naja oxiana.
Golovanov, A.P., Lomize, A.L., Arseniev, A.S., Utkin, Y.N., Tsetlin, V.I.(1993) Eur J Biochem 213: 1213-1223
- PubMed: 8504813
- DOI: https://doi.org/10.1111/j.1432-1033.1993.tb17872.x
- Primary Citation of Related Structures:
1NOR - PubMed Abstract:
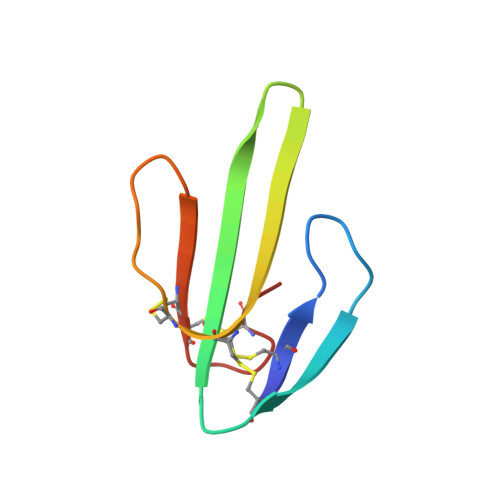
The spatial structure of neurotoxin II from the venom of the central Asian cobra Naja naja oxiana was determined by two-dimensional 1H-NMR techniques and computational analysis. Nearly complete proton resonance assignments for 61 amino acid residues have been made using two-dimensional (2D) homonuclear total correlated spectroscopy, 2D homonuclear double-quantum-filtered correlated spectroscopy and 2D homonuclear NOE spectroscopy (NOESY) experiments. The cross-peak volumes in NOESY spectra spin-spin coupling constants of vicinal protons NH-C alpha H and C alpha H-C beta H and the observation of slow deuterium exchange of amide protons were used to define local structure and a set of constraints for distance geometry program DIANA. The average root-mean-square deviations are 53 pm for backbone heavy atoms and 118 pm for all heavy atoms of 19 final neurotoxin II conformations. The spatial structure is characterized by a short double-stranded (residues 1-5 and 13-17) and a triple-stranded (residues 22-30, 33-41 and 50-54) antiparallel beta-sheets.
Organizational Affiliation:
Shemyakin Institute of Bioorganic Chemistry, Russian Academy of Sciences, Moscow.