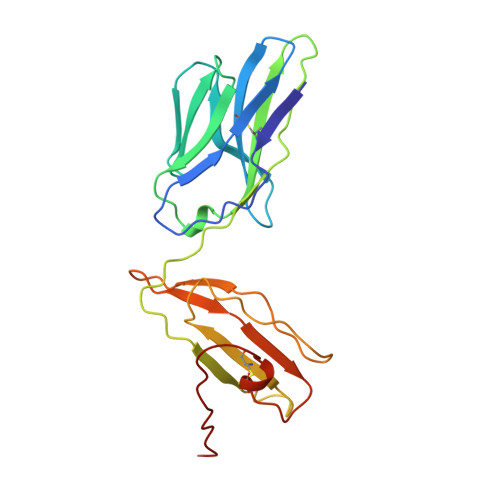
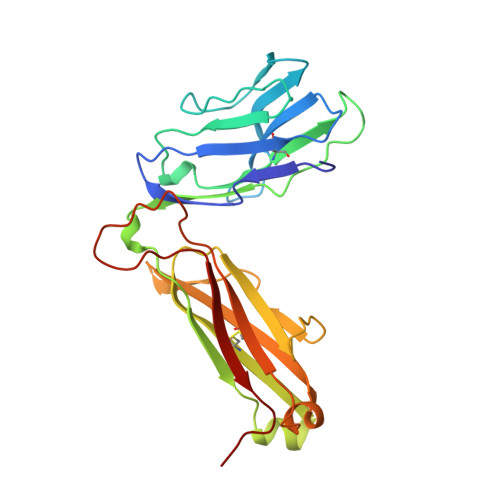
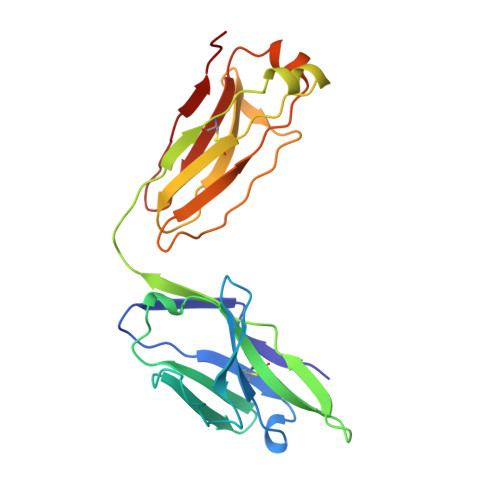
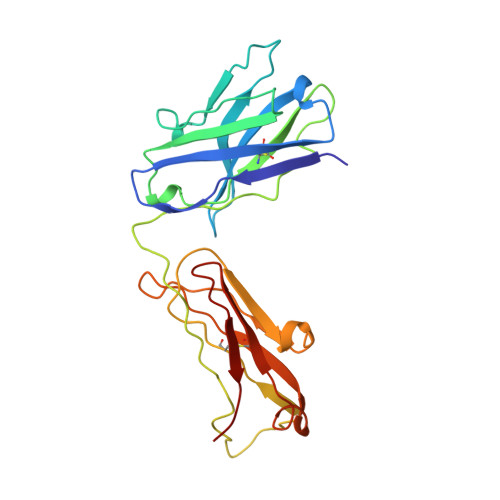
Atomic structure of an alphabeta T cell receptor (TCR) heterodimer in complex with an anti-TCR fab fragment derived from a mitogenic antibody.
Wang, J., Lim, K., Smolyar, A., Teng, M., Liu, J., Tse, A.G., Liu, J., Hussey, R.E., Chishti, Y., Thomson, C.T., Sweet, R.M., Nathenson, S.G., Chang, H.C., Sacchettini, J.C., Reinherz, E.L.(1998) EMBO J 17: 10-26
- PubMed: 9427737
- DOI: https://doi.org/10.1093/emboj/17.1.10
- Primary Citation of Related Structures:
1NFD - PubMed Abstract:
Each T cell receptor (TCR) recognizes a peptide antigen bound to a major histocompatibility complex (MHC) molecule via a clonotypic alphabeta heterodimeric structure (Ti) non-covalently associated with the monomorphic CD3 signaling components. A crystal structure of an alphabeta TCR-anti-TCR Fab complex shows an Fab fragment derived from the H57 monoclonal antibody (mAb), interacting with the elongated FG loop of the Cbeta domain, situated beneath the Vbeta domain. This loop, along with the partially exposed ABED beta sheet of Cbeta, and glycans attached to both Cbeta and Calpha domains, forms a cavity of sufficient size to accommodate a single non-glycosylated Ig domain such as the CD3epsilon ectodomain. That this asymmetrically localized site is embedded within the rigid constant domain module has implications for the mechanism of signal transduction in both TCR and pre-TCR complexes. Furthermore, quaternary structures of TCRs vary significantly even when they bind the same MHC molecule, as manifested by a unique twisting of the V module relative to the C module.
Organizational Affiliation:
Laboratory of Immunobiology, Dana-Farber Cancer Institute, Harvard Medical School, Boston, MA 02115, USA.