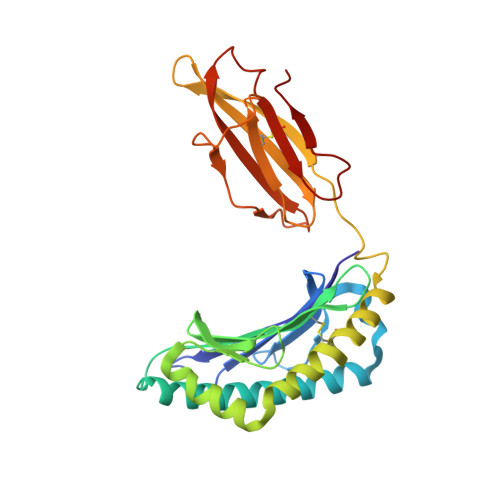
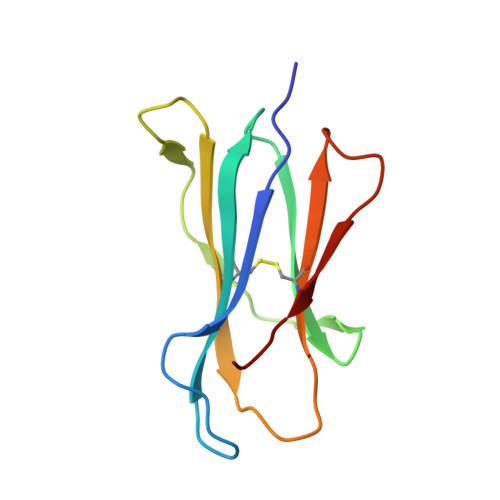
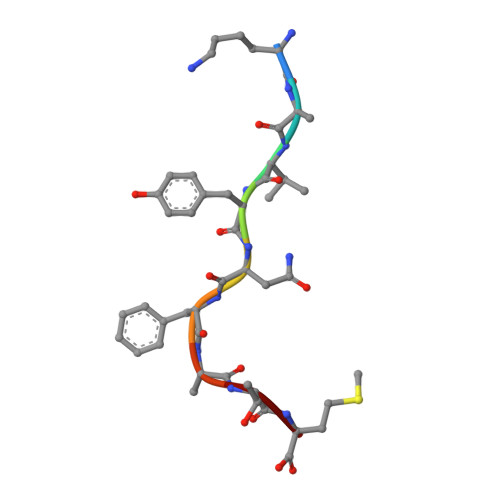
A Structural Basis for LCMV Immune Evasion. Subversion of H-2D(b) and H-2K(b) Presentation of gp33 Revealed by Comparative Crystal Structure Analyses.
Achour, A., Michaelsson, J., Harris, R.A., Odeberg, J., Grufman, P., Sandberg, J.K., Levitsky, V., Karre, K., Sandalova, T., Schneider, G.(2002) Immunity 17: 757-768
- PubMed: 12479822
- DOI: https://doi.org/10.1016/s1074-7613(02)00478-8
- Primary Citation of Related Structures:
1N59, 1N5A - PubMed Abstract:
LCMV infection of H-2(b) mice generates a CD8(+) CTL response mainly directed toward three immunodominant epitopes. One of these, gp33, is presented by both H-2D(b) and H-2K(b) MHC class I molecules. The virus can escape immune recognition in the context of both these MHC class I molecules through single mutations of the peptide. In order to understand the underlying structural mechanism, we determined the crystal structures of both complexes. The structures reveal that the peptide is presented in two diametrically opposed manners by H-2D(b) and H-2K(b), with residues used as anchor positions in one MHC class I molecule interacting with the TCR in the other. Importantly, the peptide's N-terminal residue p1K protrudes from the binding cleft in H-2K(b). We present structural evidence that explains the functional consequences of single mutations found in escape variants.
Organizational Affiliation:
Microbiology and Tumor Biology Center, Karolinska Institutet, Royal School of Technology, S-106 91 Stockholm, Sweden. adnane.achour@mtc.ki.se