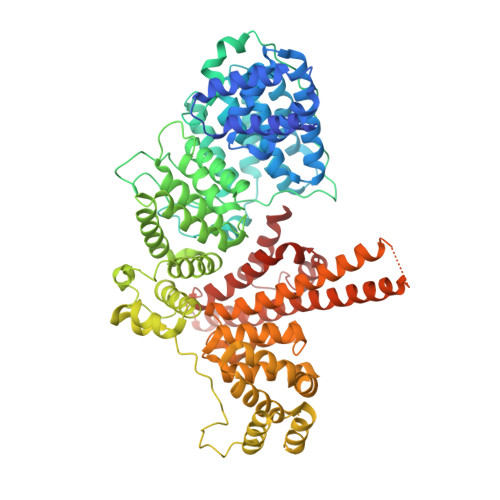
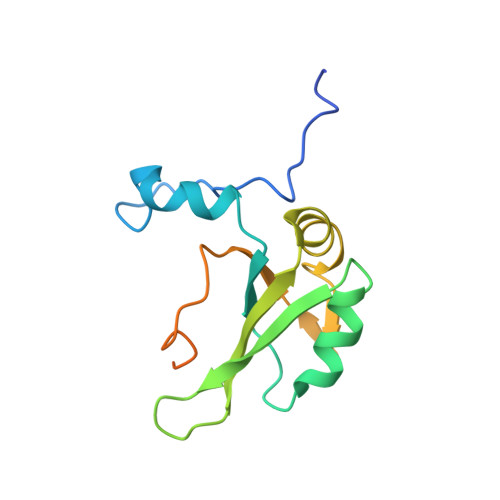
Structural basis of m7GpppG binding to the nuclear cap-binding protein complex.
Calero, G., Wilson, K., Ly, T., Rios-Steiner, J., Clardy, J., Cerione, R.(2002) Nat Struct Biol 9: 912-917
- PubMed: 12434151
- DOI: https://doi.org/10.1038/nsb874
- Primary Citation of Related Structures:
1N52, 1N54 - PubMed Abstract:
The 7-methyl guanosine cap structure of RNA is essential for key aspects of RNA processing, including pre-mRNA splicing, 3' end formation, U snRNA transport, nonsense-mediated decay and translation. Two cap-binding proteins mediate these effects: cytosolic eIF-4E and nuclear cap-binding protein complex (CBC). The latter consists of a CBP20 subunit, which binds the cap, and a CBP80 subunit, which ensures high-affinity cap binding. Here we report the 2.1 A resolution structure of human CBC with the cap analog m7GpppG, as well as the structure of unliganded CBC. Comparisons between these structures indicate that the cap induces substantial conformational changes within the N-terminal loop of CBP20, enabling Tyr 20 to join Tyr 43 in pi-pi stacking interactions with the methylated guanosine base. CBP80 stabilizes the movement of the N-terminal loop of CBP20 and locks the CBC into a high affinity cap-binding state. The structure for the CBC bound to m7GpppG highlights interesting similarities and differences between CBC and eIF-4E, and provides insights into the regulatory mechanisms used by growth factors and other extracellular stimuli to influence the cap-binding state of the CBC.
Organizational Affiliation:
Department of Chemistry and Chemical Biology, Baker Laboratory, Cornell University, Ithaca, New York 14853, USA.