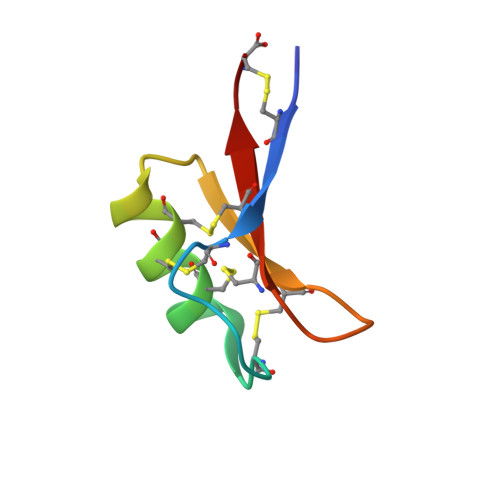
Structure of Petunia hybrida defensin 1, a novel plant defensin with five disulfide bonds
Janssen, B.J.C., Schirra, H.J., Lay, F.T., Anderson, M.A., Craik, D.J.(2003) Biochemistry 42: 8214-8222
- PubMed: 12846570
- DOI: https://doi.org/10.1021/bi034379o
- Primary Citation of Related Structures:
1N4N - PubMed Abstract:
The structure of a novel plant defensin isolated from the flowers of Petunia hybrida has been determined by (1)H NMR spectroscopy. P. hybrida defensin 1 (PhD1) is a basic, cysteine-rich, antifungal protein of 47 residues and is the first example of a new subclass of plant defensins with five disulfide bonds whose structure has been determined. PhD1 has the fold of the cysteine-stabilized alphabeta motif, consisting of an alpha-helix and a triple-stranded antiparallel beta-sheet, except that it contains a fifth disulfide bond from the first loop to the alpha-helix. The additional disulfide bond is accommodated in PhD1 without any alteration of its tertiary structure with respect to other plant defensins. Comparison of its structure with those of classic, four-disulfide defensins has allowed us to identify a previously unrecognized hydrogen bond network that is integral to structure stabilization in the family.
Organizational Affiliation:
Institute for Molecular Bioscience, ARC Special Research Center for Functional and Applied Genomics, The University of Queensland, Brisbane, Queensland 4072, Australia.