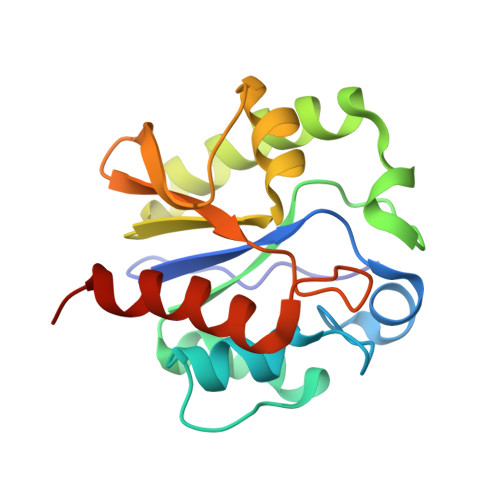
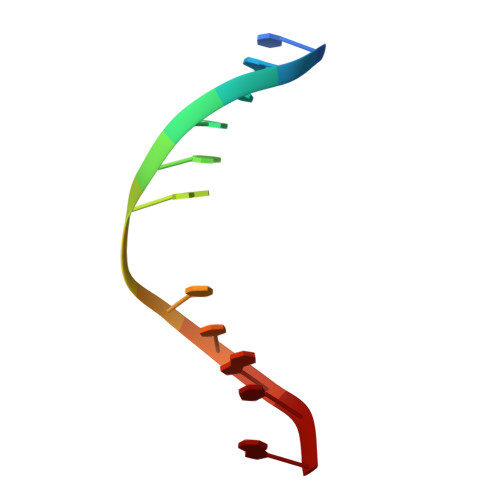
Crystal structure of a G:T/U mismatch-specific DNA glycosylase: mismatch recognition by complementary-strand interactions.
Barrett, T.E., Savva, R., Panayotou, G., Barlow, T., Brown, T., Jiricny, J., Pearl, L.H.(1998) Cell 92: 117-129
- PubMed: 9489705
- DOI: https://doi.org/10.1016/s0092-8674(00)80904-6
- Primary Citation of Related Structures:
1MUG, 1MWI - PubMed Abstract:
G:U mismatches resulting from deamination of cytosine are the most common promutagenic lesions occurring in DNA. Uracil is removed in a base-excision repair pathway by uracil DNA-glycosylase (UDG), which excises uracil from both single- and double-stranded DNA. Recently, a biochemically distinct family of DNA repair enzymes has been identified, which excises both uracil and thymine, but only from mispairs with guanine. Crystal structures of the mismatch-specific uracil DNA-glycosylase (MUG) from E. coli, and of a DNA complex, reveal a remarkable structural and functional homology to UDGs despite low sequence identity. Details of the MUG structure explain its thymine DNA-glycosylase activity and the specificity for G:U/T mispairs, which derives from direct recognition of guanine on the complementary strand.
Organizational Affiliation:
Department of Biochemistry and Molecular Biology, University College London, United Kingdom.