Noncompetitive antibody neutralization of IL-10 revealed by protein engineering and x-ray crystallography.
Josephson, K., Jones, B.C., Walter, L.J., DiGiacomo, R., Indelicato, S.R., Walter, M.R.(2002) Structure 10: 981-987
- PubMed: 12121653
- DOI: https://doi.org/10.1016/s0969-2126(02)00791-8
- Primary Citation of Related Structures:
1LK3 - PubMed Abstract:
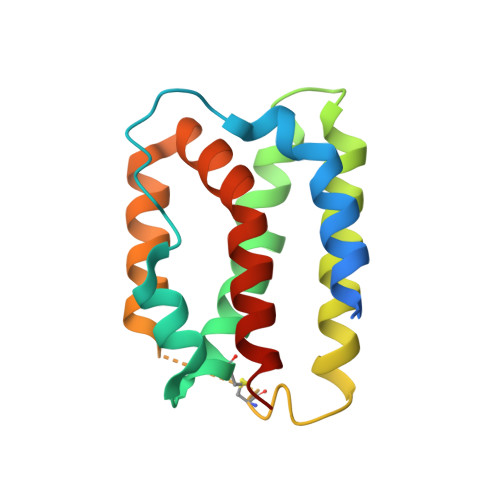
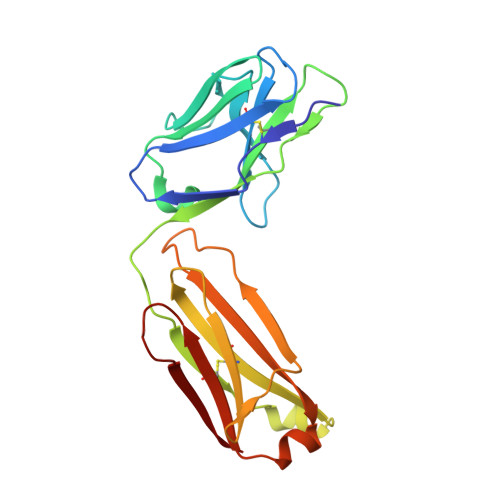
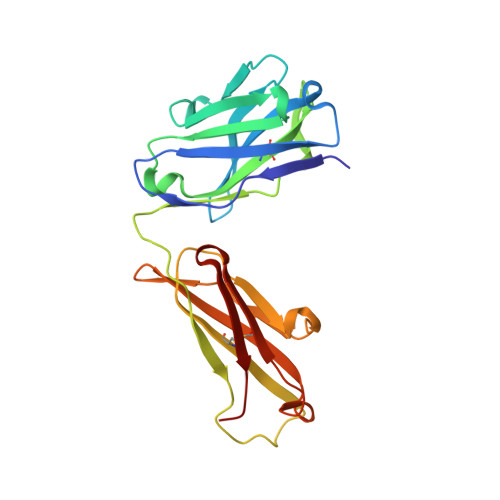
IL-10 is a dimeric cytokine that must engage its high-affinity cell surface receptor, IL-10R1, to induce multiple cellular activities. Here we report the 1.9 A crystal structure of an engineered IL-10 monomer (IL-10M1) in complex with a neutralizing Fab fragment (9D7Fab). 9D7Fab and IL-10R1 bind distinct nonoverlapping surfaces on IL-10M1. Antagonism of the IL-10M1/IL-10R1 interaction is the result of 9D7Fab-induced conformational changes in the CD loop of IL-10M1 that indirectly alter the structure of the IL-10R1 binding site. A single mutation (Ile87Ala) in the same CD loop region of the Epstein-Barr virus IL-10 (ebvIL-10) also reduces IL-10R1 binding affinity, suggesting that ebvIL-10 and 9D7Fab use similar allosteric mechanisms to modulate IL-10R1 affinity and biological activity.
Organizational Affiliation:
Center for Biophysical Sciences and Engineering, University of Alabama, Birmingham, 35294, USA.