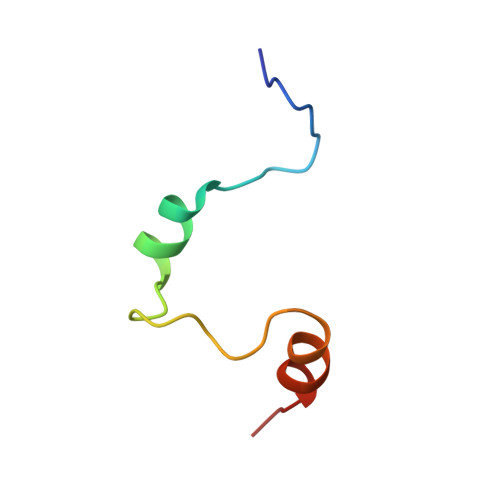
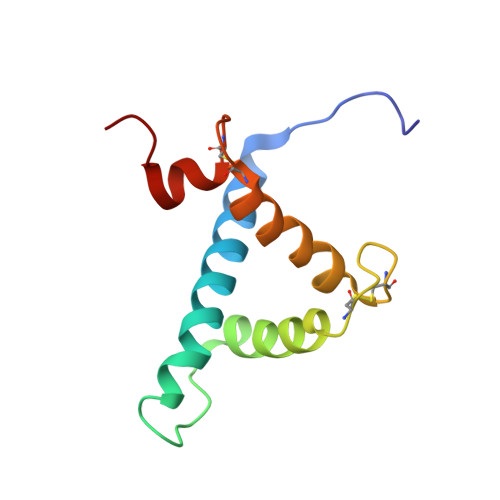
Structural basis for recruitment of CBP/p300 by hypoxia-inducible factor-1 alpha.
Freedman, S.J., Sun, Z.Y., Poy, F., Kung, A.L., Livingston, D.M., Wagner, G., Eck, M.J.(2002) Proc Natl Acad Sci U S A 99: 5367-5372
- PubMed: 11959990
- DOI: https://doi.org/10.1073/pnas.082117899
- Primary Citation of Related Structures:
1L3E - PubMed Abstract:
Adaptation to hypoxia is mediated by transactivation of hypoxia-responsive genes by hypoxia-inducible factor-1 (HIF-1) in complex with the CBP and p300 transcriptional coactivators. We report the solution structure of the cysteine/histidine-rich 1 (CH1) domain of p300 bound to the C-terminal transactivation domain of HIF-1 alpha. CH1 has a triangular geometry composed of four alpha-helices with three intervening Zn(2+)-coordinating centers. CH1 serves as a scaffold for folding of the HIF-1 alpha C-terminal transactivation domain, which forms a vise-like clamp on the CH1 domain that is stabilized by extensive hydrophobic and polar interactions. The structure reveals the mechanism of specific recognition of p300 by HIF-1 alpha, and shows how HIF-1 alpha transactivation is regulated by asparagine hydroxylation.
Organizational Affiliation:
Division of Hematology/Oncology, Beth Israel Deaconess Medical Center, 330 Brookline Avenue, Boston, MA 02215, USA.