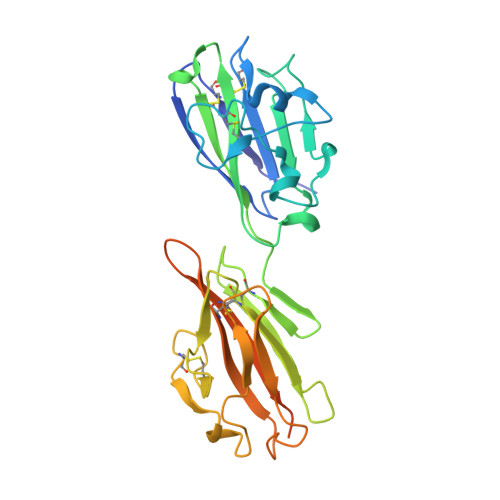
Structure of the immunodominant surface antigen from the Toxoplasma gondii SRS superfamily.
He, X.L., Grigg, M.E., Boothroyd, J.C., Garcia, K.C.(2002) Nat Struct Biol 9: 606-611
- PubMed: 12091874
- DOI: https://doi.org/10.1038/nsb819
- Primary Citation of Related Structures:
1KZQ - PubMed Abstract:
Toxoplasma gondii is a persistent protozoan parasite capable of infecting almost any warm-blooded vertebrate. The surface of Toxoplasma is coated with a family of developmentally regulated glycosylphosphatidylinositol (GPI)-linked proteins (SRSs), of which SAG1 is the prototypic member. SRS proteins mediate attachment to host cells and interface with the host immune response to regulate the virulence of the parasite. The 1.7 A structure of the immunodominant SAG1 antigen reveals a homodimeric configuration in which the dimeric interface is mediated by an extended beta-sheet that forms a deep groove lined with positively charged amino acids. This basic groove seems to be conserved among SRS proteins and potentially serves as a sulfated proteoglycan-binding site on target cell surfaces, thus rationalizing the promiscuous attachment properties of Toxoplasma to a broad range of host cell types.
Organizational Affiliation:
Department of Microbiology and Immunology, Stanford University School of Medicine, Stanford, Cailfornia 94305-5124, USA.