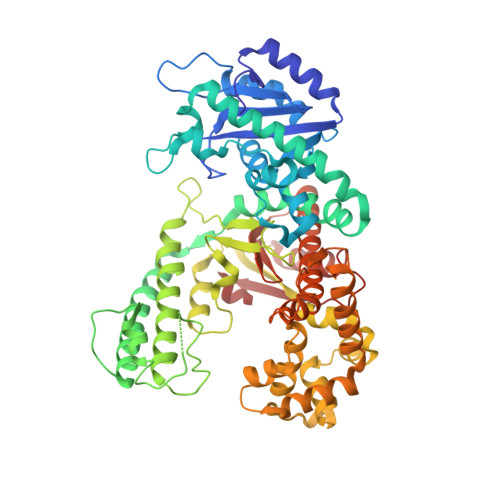
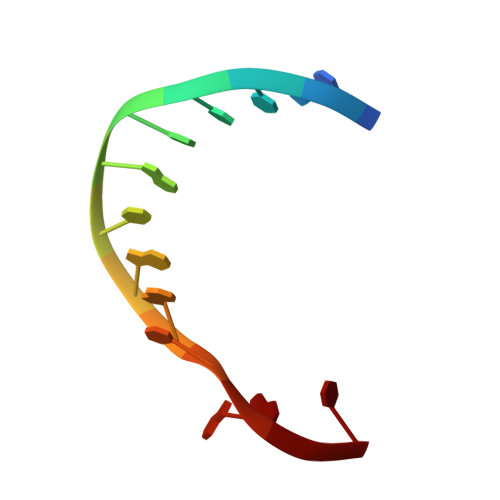
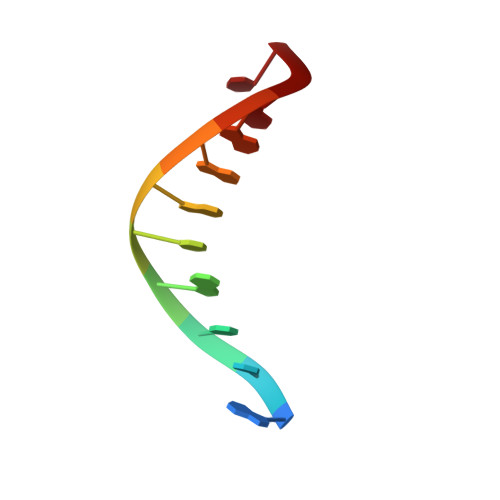
Structure of DNA polymerase I Klenow fragment bound to duplex DNA.
Beese, L.S., Derbyshire, V., Steitz, T.A.(1993) Science 260: 352-355
- PubMed: 8469987
- DOI: https://doi.org/10.1126/science.8469987
- Primary Citation of Related Structures:
1KLN - PubMed Abstract:
Klenow fragment of Escherichia coli DNA polymerase I, which was cocrystallized with duplex DNA, positioned 11 base pairs of DNA in a groove that lies at right angles to the cleft that contains the polymerase active site and is adjacent to the 3' to 5' exonuclease domain. When the fragment bound DNA, a region previously referred to as the "disordered domain" became more ordered and moved along with two helices toward the 3' to 5' exonuclease domain to form the binding groove. A single-stranded, 3' extension of three nucleotides bound to the 3' to 5' exonuclease active site. Although this cocrystal structure appears to be an editing complex, it suggests that the primer strand approaches the catalytic site of the polymerase from the direction of the 3' to 5' exonuclease domain and that the duplex DNA product may bend to enter the cleft that contains the polymerase catalytic site.
Organizational Affiliation:
Department of Molecular Biophysics and Biochemistry, Yale University, New Haven, CT 06511.