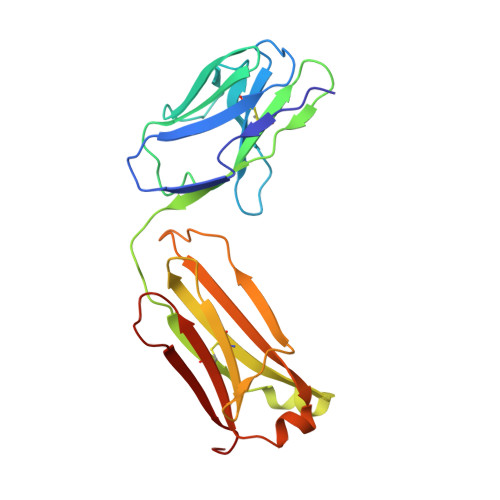
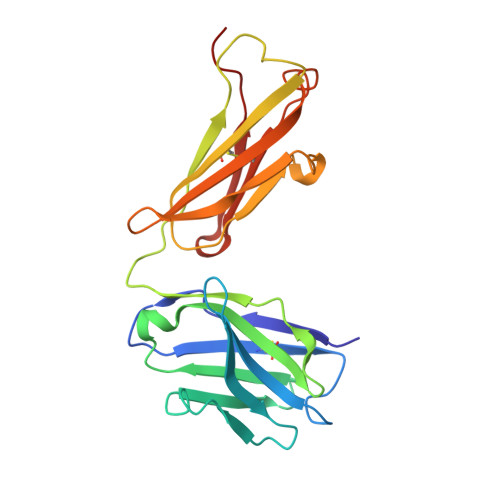
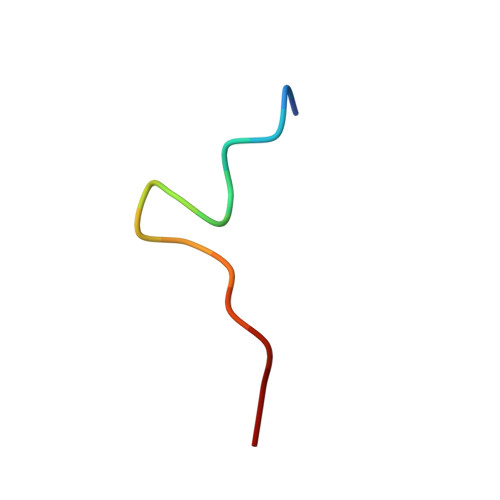
Crystal structure of an antibody bound to an immunodominant peptide epitope: novel features in peptide-antibody recognition.
Nair, D.T., Singh, K., Sahu, N., Rao, K.V., Salunke, D.M.(2000) J Immunol 165: 6949-6955
- PubMed: 11120821
- DOI: https://doi.org/10.4049/jimmunol.165.12.6949
- Primary Citation of Related Structures:
1KCR - PubMed Abstract:
The crystal structure of Fab of an Ab PC283 complexed with its corresponding peptide Ag, PS1 (HQLDPAFGANSTNPD), derived from the hepatitis B virus surface Ag was determined. The PS1 stretch Gln2P to Phe7P is present in the Ag binding site of the Ab, while the next three residues of the peptide are raised above the binding groove. The residues Ser11P, Thr12P, and Asn13P then loop back onto the Ag-binding site of the Ab. The last two residues, Pro14P and Asp15P, extend outside the binding site without forming any contacts with the Ab. The PC283-PS1 complex is among the few examples where the light chain complementarity-determining regions show more interactions than the heavy chain complementarity-determining regions, and a distal framework residue is involved in Ag binding. As seen from the crystal structure, most of the contacts between peptide and Ab are through the five residues, Leu3-Asp4-Pro5-Ala6-Phe7, of PS1. The paratope is predominantly hydrophobic with aromatic residues lining the binding pocket, although a salt bridge also contributes to stabilizing the Ag-Ab interaction. The molecular surface area buried upon PS1 binding is 756 A(2) for the peptide and 625 A(2) for the Fab, which is higher than what has been seen to date for Ab-peptide complexes. A comparison between PC283 structure and a homology model of its germline ancestor suggests that paratope optimization for PS1 occurs by improving both charge and shape complementarity.
Organizational Affiliation:
National Institute of Immunology and International Center of Genetic Engineering and Biotechnology, New Delhi, India.