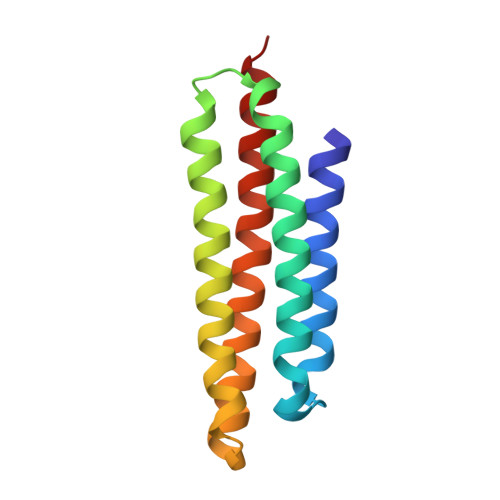
The focal adhesion targeting (FAT) region of focal adhesion kinase is a four-helix bundle that binds paxillin
Hayashi, I., Vuori, K., Liddington, R.C.(2002) Nat Struct Biol 9: 101-106
- PubMed: 11799401
- DOI: https://doi.org/10.1038/nsb755
- Primary Citation of Related Structures:
1K40 - PubMed Abstract:
Focal adhesion kinase (FAK) is a tyrosine kinase found in focal adhesions, intracellular signaling complexes that are formed following engagement of the extracellular matrix by integrins. The C-terminal 'focal adhesion targeting' (FAT) region is necessary and sufficient for localizing FAK to focal adhesions. We have determined the crystal structure of FAT and show that it forms a four-helix bundle that resembles those found in two other proteins involved in cell adhesion, alpha-catenin and vinculin. The binding of FAT to the focal adhesion protein, paxillin, requires the integrity of the helical bundle, whereas binding to another focal adhesion protein, talin, does not. We show by mutagenesis that paxillin binding involves two hydrophobic patches on opposite faces of the bundle and propose a model in which two LD motifs of paxillin adopt amphipathic helices that augment the hydrophobic core of FAT, creating a six-helix bundle.
Organizational Affiliation:
Program on Cell Adhesion, The Burnham Institute, 10901 North Torrey Pines Road, La Jolla, California 92037,USA.