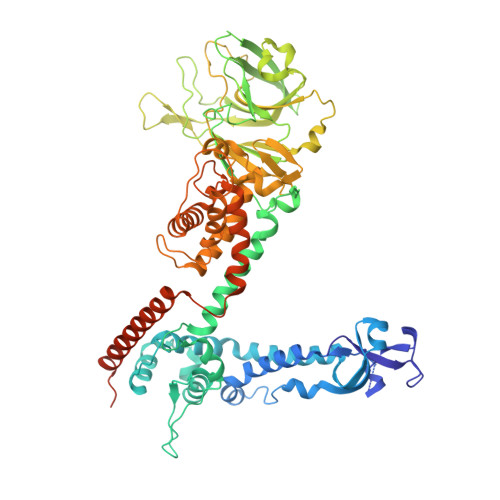
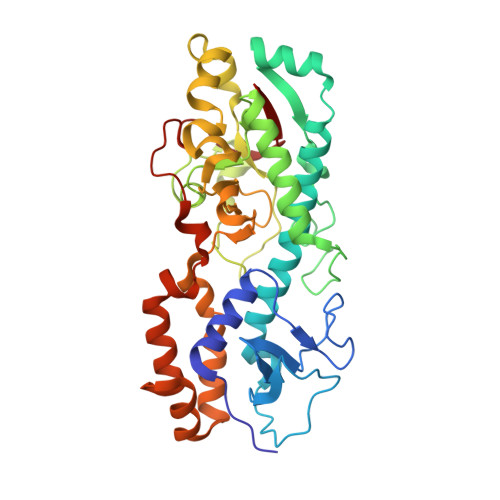
Structure of the reovirus membrane-penetration protein, Mu1, in a complex with is protector protein, Sigma3.
Liemann, S., Chandran, K., Baker, T.S., Nibert, M.L., Harrison, S.C.(2002) Cell 108: 283-295
- PubMed: 11832217
- DOI: https://doi.org/10.1016/s0092-8674(02)00612-8
- Primary Citation of Related Structures:
1JMU - PubMed Abstract:
Cell entry by nonenveloped animal viruses requires membrane penetration without membrane fusion. The reovirus penetration agent is the outer-capsid protein, Mu1. The structure of Mu1, complexed with its "protector" protein, Sigma3, and the fit of this Mu1(3)Sigma3(3) heterohexameric complex into the cryoEM image of an intact virion, reveal molecular events essential for viral penetration. Autolytic cleavage divides Mu1 into myristoylated Mu1N and Mu1C. A long hydrophobic pocket can receive the myristoyl group. Dissociation of Mu1N, linked to a major conformational change of the entire Mu1 trimer, must precede myristoyl-group insertion into the cellular membrane. A myristoyl switch, coupling exposure of the fatty acid chain, autolytic cleavage of Mu1N, and long-range molecular rearrangement of Mu1C, thus appears to be part of the penetration mechanism.
Organizational Affiliation:
Howard Hughes Medical Institute, Children's Hospital, Harvard Medical School, 320 Longwood Avenue, Boston, MA 02115, USA.