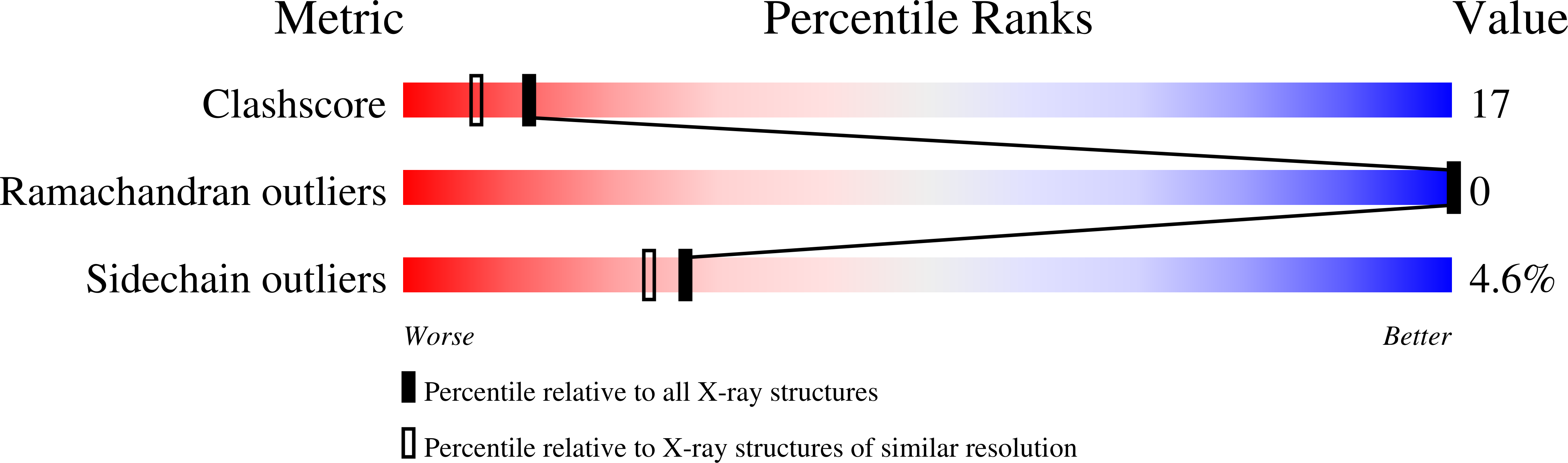NTF2 monomer-dimer equilibrium.
Chaillan-Huntington, C., Butler, P.J., Huntington, J.A., Akin, D., Feldherr, C., Stewart, M.(2001) J Mol Biol 314: 465-477
- PubMed: 11846560
- DOI: https://doi.org/10.1006/jmbi.2001.5136
- Primary Citation of Related Structures:
1JB2, 1JB4, 1JB5 - PubMed Abstract:
Nuclear transport factor 2 (NTF2) mediates nuclear import of RanGDP, a central component of many nuclear trafficking pathways. NTF2 is a homodimer and each chain has independent binding sites for RanGDP and nuclear pore proteins (nucleoporins) that contain FxFG sequence repeats. We show here that the monomer-dimer dissociation constant for NTF2 obtained by sedimentation equilibrium ultracentrifugation is in the micromolar range, indicating that a substantial proportion of cellular NTF2 may be monomeric. To investigate the functional significance of NTF2 dimerization, we engineered a series of point mutations at the dimerization interface and one of these (M118E) remained monomeric below concentrations of 150 microM. CD spectra and X-ray crystallography showed that M118E-NTF2 preserved the wild-type NTF2 fold, although its thermal stability was 20 deg. C lower than that of the wild-type. M118E-NTF2 bound both RanGDP and FxFG nucleoporins less strongly, suggesting that dissociation of the NTF2 dimer could facilitate RanGDP release and thus nucleotide exchange after it had been transported into the nucleus. Moreover, colloidal gold coated with M118E-NTF2 showed reduced binding to Xenopus oocyte nuclear pores. Overall, our results indicate that dimer formation is important for NTF2 function and give insight into the formation of heterodimers by mRNA export factors such as TAP1 and NXT1 that contain NTF2-homology domains.
Organizational Affiliation:
Laboratory of Molecular Biology, MRC, Hills Road, Cambridge, CB2 2QH, England.