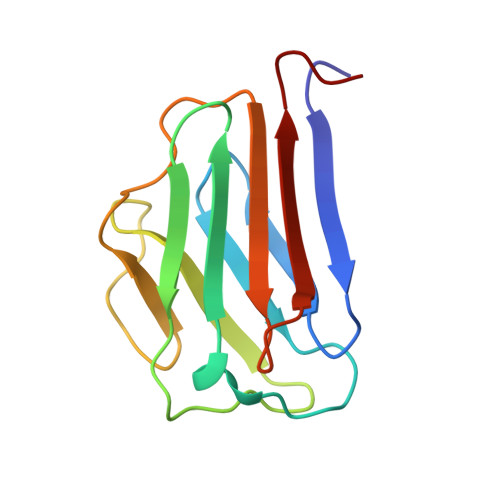
The Ig-like structure of the C-terminal domain of lamin A/C, mutated in muscular dystrophies, cardiomyopathy, and partial lipodystrophy.
Krimm, I., Ostlund, C., Gilquin, B., Couprie, J., Hossenlopp, P., Mornon, J.P., Bonne, G., Courvalin, J.C., Worman, H.J., Zinn-Justin, S.(2002) Structure 10: 811-823
- PubMed: 12057196
- DOI: https://doi.org/10.1016/s0969-2126(02)00777-3
- Primary Citation of Related Structures:
1IVT - PubMed Abstract:
Lamins are nuclear intermediate filaments that, together with lamin-associated proteins, maintain nuclear shape and provide a structural support for chromosomes and replicating DNA. We have determined the solution structure of the human lamin A/C C-terminal globular domain which contains specific mutations causing four different heritable diseases. This domain encompasses residues 430-545 and adopts an Ig-like fold of type s. We have also characterized by NMR and circular dichroism the structure and thermostability of three mutants, R453W and R482W/Q, corresponding to "hot spots" causing Emery-Dreifuss muscular dystrophy and Dunnigan-type lipodystrophy, respectively. Our structure determination and mutant analyses clearly show that the consequences of the mutations causing muscle-specific diseases or lipodystrophy are different at the molecular level.
Organizational Affiliation:
Département d'Ingénierie et d'Etudes des Protéines, CEA Saclay, 91191, Gif-sur-Yvette, France.