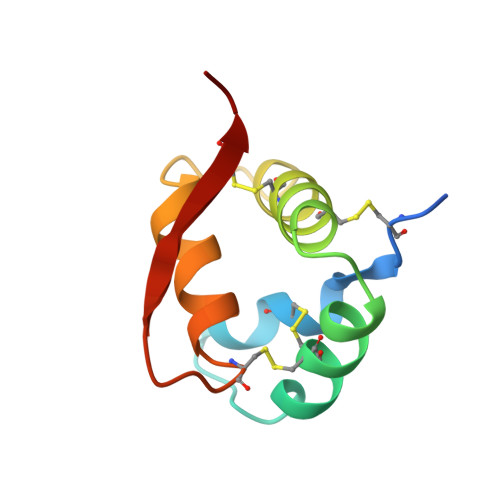
Crystal structure of hydrophobic protein from soybean; a member of a new cysteine-rich family.
Baud, F., Pebay-Peyroula, E., Cohen-Addad, C., Odani, S., Lehmann, M.S.(1993) J Mol Biol 231: 877-887
- PubMed: 8515457
- DOI: https://doi.org/10.1006/jmbi.1993.1334
- Primary Citation of Related Structures:
1HYP - PubMed Abstract:
X-ray diffraction methods have been used to determine the structure of the 8.3 kDa hydrophobic protein from soybean and to refine the atomic co-ordinates to a crystallographic R-factor of 18.7% at 1.8 A resolution. The molecule is a four-helix bundle, which together with the connecting loops and a twisted beta-strand form a spiral. The surface contains 70% apolar atoms, and the crystal packing is dominated by hydrophobic interactions, producing a two-dimensional sheet of protein molecules. Most of the 59 water molecules located are involved in hydrophilic contacts and their structural organization does not seem to be affected by the high hydrophobicity of the molecule. From the protein fold it appears that three of the four disulphide bridges are important for keeping the amino and carboxyl-terminal segments in place in the native form, while the central part of the molecule is stabilized by many hydrophobic interactions. Although the protein function is not known, a number of possibilities can be excluded on experimental grounds and by comparison with other members of the family.
Organizational Affiliation:
Institut Laue-Langevin, Grenoble, France.