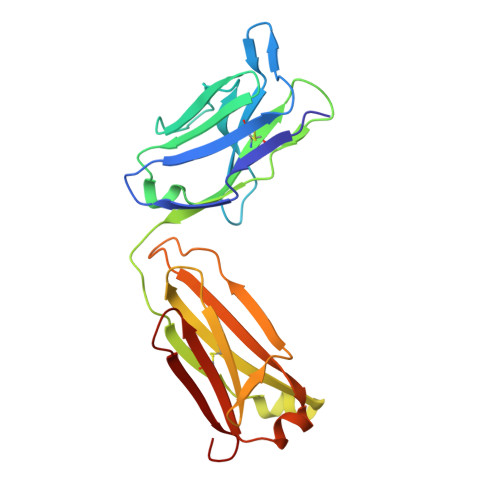
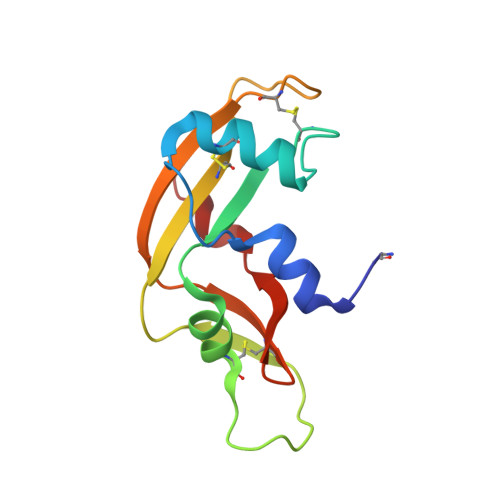
The Crystal Structure of Human Angiogenin in Complex with an Antitumor Neutralizing Antibody
Chavali, G.B., Papageorgiou, A.C., Olson, K., Fett, J., Hu, G., Shapiro, R., Acharya, K.R.(2003) Structure 11: 875
- PubMed: 12842050
- DOI: https://doi.org/10.1016/s0969-2126(03)00131-x
- Primary Citation of Related Structures:
1H0D - PubMed Abstract:
The murine monoclonal antibody 26-2F neutralizes the angiogenic and ribonucleolytic activities of human angiogenin (ANG) and is highly effective in preventing the establishment and metastatic dissemination of human tumors in athymic mice. Here we report a 2.0 A resolution crystal structure for the complex of ANG with the Fab fragment of 26-2F that reveals the detailed interactions between ANG and the complementarity-determining regions (CDRs) of the antibody. Surprisingly, Fab binding induces a dramatic conformational change in the cell binding region of ANG at the opposite end of the molecule from the combining site; crosslinking experiments indicate that this rearrangement also occurs in solution. The ANG-Fab complex structure should be invaluable for designing maximally humanized versions of 26-2F for potential clinical use.
Organizational Affiliation:
Department of Biology and Biochemistry, University of Bath, Claverton Down, BA2 7AY, Bath, United Kingdom.