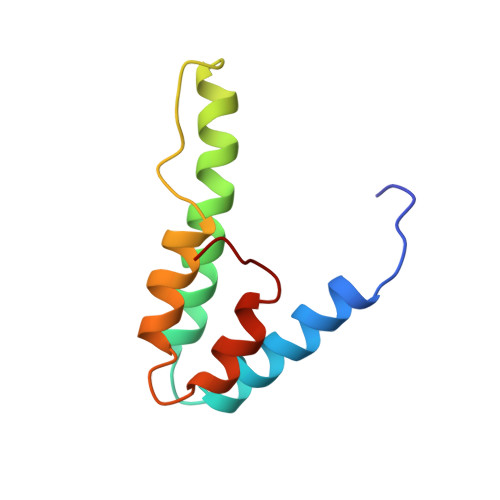
Solution structure of the interacting domains of the Mad-Sin3 complex: implications for recruitment of a chromatin-modifying complex.
Brubaker, K., Cowley, S.M., Huang, K., Loo, L., Yochum, G.S., Ayer, D.E., Eisenman, R.N., Radhakrishnan, I.(2000) Cell 103: 655-665
- PubMed: 11106735
- DOI: https://doi.org/10.1016/s0092-8674(00)00168-9
- Primary Citation of Related Structures:
1G1E - PubMed Abstract:
Gene-specific targeting of the Sin3 corepressor complex by DNA-bound repressors is an important mechanism of gene silencing in eukaryotes. The Sin3 corepressor specifically associates with a diverse group of transcriptional repressors, including members of the Mad family, that play crucial roles in development. The NMR structure of the complex formed by the PAH2 domain of mammalian Sin3A with the transrepression domain (SID) of human Mad1 reveals that both domains undergo mutual folding transitions upon complex formation generating an unusual left-handed four-helix bundle structure and an amphipathic alpha helix, respectively. The SID helix is wedged within a deep hydrophobic pocket defined by two PAH2 helices. Structure-function analyses of the Mad-Sin3 complex provide a basis for understanding the underlying mechanism(s) that lead to gene silencing.
Organizational Affiliation:
Department of Biochemistry, Molecular Biology, and Cell Biology, Northwestern University, Evanston, IL 60208, USA.