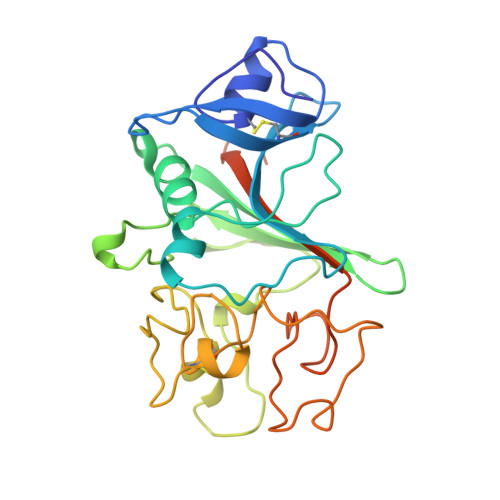
Crystal structure of a 30 kDa C-terminal fragment from the gamma chain of human fibrinogen.
Yee, V.C., Pratt, K.P., Cote, H.C., Trong, I.L., Chung, D.W., Davie, E.W., Stenkamp, R.E., Teller, D.C.(1997) Structure 5: 125-138
- PubMed: 9016719
- DOI: https://doi.org/10.1016/s0969-2126(97)00171-8
- Primary Citation of Related Structures:
1FIB, 1FIC, 1FID - PubMed Abstract:
Blood coagulation occurs by a cascade of zymogen activation resulting from minor proteolysis. The final stage of coagulation involves thrombin generation and limited proteolysis of fibrinogen to give spontaneously polymerizing fibrin. The resulting fibrin network is covalently crosslinked by factor XIIIa to yield a stable blood clot. Fibrinogen is a 340 kDa glycoprotein composed of six polypeptide chains, (alphabetagamma)2, held together by 29 disulfide bonds. The globular C terminus of the gamma chain contains a fibrin-polymerization surface, the principal factor XIIIa crosslinking site, the platelet receptor recognition site, and a calcium-binding site. Structural information on this domain should thus prove helpful in understanding clot formation.
Organizational Affiliation:
Department of Biochemistry, Biomolecular Structure Center, University of Washington, Seattle, WA 98195, USA.