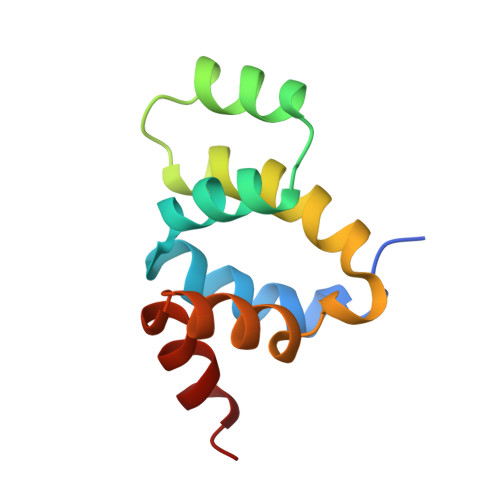
The solution structure of FADD death domain. Structural basis of death domain interactions of Fas and FADD.
Jeong, E.J., Bang, S., Lee, T.H., Park, Y.I., Sim, W.S., Kim, K.S.(1999) J Biol Chem 274: 16337-16342
- PubMed: 10347191
- DOI: https://doi.org/10.1074/jbc.274.23.16337
- Primary Citation of Related Structures:
1FAD - PubMed Abstract:
A signal of Fas-mediated apoptosis is transferred through an adaptor protein Fas-associated death domain protein (FADD) by interactions between the death domains of Fas and FADD. To understand the signal transduction mechanism of Fas-mediated apoptosis, we solved the solution structure of a murine FADD death domain. It consists of six helices arranged in a similar fold to the other death domains. The interactions between the death domains of Fas and FADD analyzed by site-directed mutagenesis indicate that charged residues in helices alpha2 and alpha3 are involved in death domain interactions, and the interacting helices appear to interact in anti-parallel pattern, alpha2 of FADD with alpha3 of Fas and vice versa.
Organizational Affiliation:
Structural Biology Center, Korea Institute of Science and Technology, Seoul, 130-650, Korea University, Seoul, 136-701, Korea.