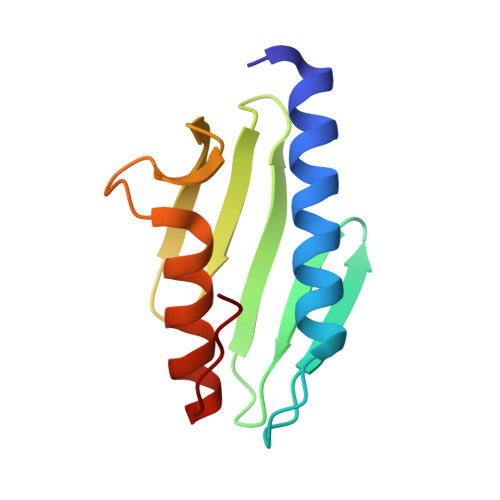
Crystal structure of Escherichia coli CyaY protein reveals a previously unidentified fold for the evolutionarily conserved frataxin family.
Cho, S.J., Lee, M.G., Yang, J.K., Lee, J.Y., Song, H.K., Suh, S.W.(2000) Proc Natl Acad Sci U S A 97: 8932-8937
- PubMed: 10908679
- DOI: https://doi.org/10.1073/pnas.160270897
- Primary Citation of Related Structures:
1EW4 - PubMed Abstract:
Friedreich ataxia is an autosomal recessive neurodegenerative disease caused by defects in the FRDA gene, which encodes a mitochondrial protein called frataxin. Frataxin is evolutionarily conserved, with homologs identified in mammals, worms, yeast, and bacteria. The CyaY proteins of gamma-purple bacteria are believed to be closely related to the ancestor of frataxin. In this study, we have determined the crystal structure of the CyaY protein from Escherichia coli at 1.4-A resolution. It reveals a protein fold consisting of a six-stranded antiparallel beta-sheet flanked on one side by two alpha-helices. This fold is likely to be shared by all members of the conserved frataxin family. This study also provides a framework for the interpretation of disease-associated mutations in frataxin and for understanding the possible functions of this protein family.
Organizational Affiliation:
Department of Chemistry, Seoul National University, Seoul 151-742, Korea.