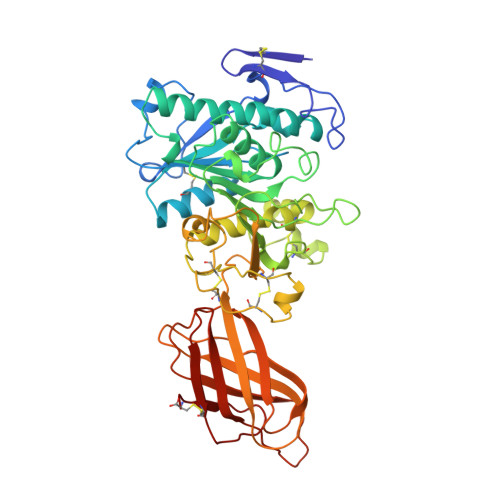
Lipase activation by nonionic detergents. The crystal structure of the porcine lipase-colipase-tetraethylene glycol monooctyl ether complex.
Hermoso, J., Pignol, D., Kerfelec, B., Crenon, I., Chapus, C., Fontecilla-Camps, J.C.(1996) J Biol Chem 271: 18007-18016
- PubMed: 8663362
- DOI: https://doi.org/10.1074/jbc.271.30.18007
- Primary Citation of Related Structures:
1ETH - PubMed Abstract:
The crystal structure of the ternary porcine lipase-colipase-tetra ethylene glycol monooctyl ether (TGME) complex has been determined at 2.8 A resolution. The crystals belong to the cubic space group F23 with a = 289.1 A and display a strong pseudo-symmetry corresponding to a P23 lattice. Unexpectedly, the crystalline two-domain lipase is found in its open configuration. This indicates that in the presence of colipase, pure micelles of the nonionic detergent TGME are able to activate the enzyme; a process that includes the movement of an N-terminal domain loop (the flap). The effects of TGME and colipase have been confirmed by chemical modification of the active site serine residue using diisopropyl p-nitrophenylphosphate (E600). In addition, the presence of a TGME molecule tightly bound to the active site pocket shows that TGME acts as a substrate analog, thus possibly explaining the inhibitory effect of this nonionic detergent on emulsified substrate hydrolysis at submicellar concentrations. A comparison of the lipase-colipase interactions between our porcine complex and the human-porcine complex (van Tilbeurgh, H., Egloff, M.-P., Martinez, C., Rugani, N., Verger, R., and Cambillau, C.(1993) Nature 362, 814-820) indicates that except for one salt bridge interaction, they are conserved. Analysis of the superimposed complexes shows a 5.4 degrees rotation on the relative position of the N-terminal domains excepting the flap that moves in a concerted fashion with the C-terminal domain. This flexibility may be important for the binding of the complex to the water-lipid interface.
Organizational Affiliation:
Laboratoire de Cristallographie et de Cristallogénèse des Protéines, Institut de Biologie Structurale Jean-Pierre Ebel, CEA-CNRS, Grenoble, France.