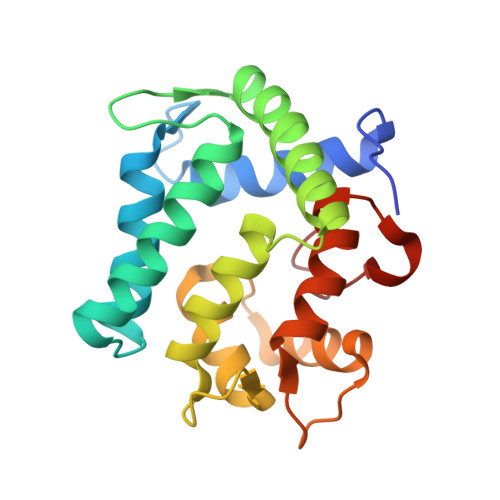
The crystal structure of the photoprotein aequorin at 2.3 A resolution.
Head, J.F., Inouye, S., Teranishi, K., Shimomura, O.(2000) Nature 405: 372-376
- PubMed: 10830969
- DOI: https://doi.org/10.1038/35012659
- Primary Citation of Related Structures:
1EJ3 - PubMed Abstract:
Aequorin is a calcium-sensitive photoprotein originally obtained from the jellyfish Aequorea aequorea. Because it has a high sensitivity to calcium ions and is biologically harmless, aequorin is widely used as a probe to monitor intracellular levels of free calcium. The aequorin molecule contains four helix-loop-helix 'EF-hand' domains, of which three can bind calcium. The molecule also contains coelenterazine as its chromophoric ligand. When calcium is added, the protein complex decomposes into apoaequorin, coelenteramide and CO2, accompanied by the emission of light. Apoaequorin can be regenerated into active aequorin in the absence of calcium by incubation with coelenterazine, oxygen and a thiol agent. Cloning and expression of the complementary DNA for aequorin were first reported in 1985 (refs 2, 6), and growth of crystals of the recombinant protein has been described; however, techniques have only recently been developed to prepare recombinant aequorin of the highest purity, permitting a full crystallographic study. Here we report the structure of recombinant aequorin determined by X-ray crystallography. Aequorin is found to be a globular molecule containing a hydrophobic core cavity that accommodates the ligand coelenterazine-2-hydroperoxide. The structure shows protein components stabilizing the peroxide and suggests a mechanism by which calcium activation may occur.
Organizational Affiliation:
Department of Physiology, Boston University School of Medicine, Massachusetts 02118, USA. jfh@medxtal.bu.edu