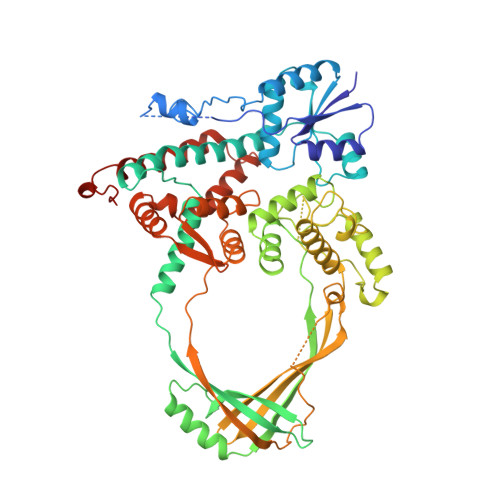
Three-dimensional structure of the 67K N-terminal fragment of E. coli DNA topoisomerase I.
Lima, C.D., Wang, J.C., Mondragon, A.(1994) Nature 367: 138-146
- PubMed: 8114910
- DOI: https://doi.org/10.1038/367138a0
- Primary Citation of Related Structures:
1ECL - PubMed Abstract:
The three-dimensional structure of the 67K amino-terminal fragment of Escherichia coli DNA topoisomerase I has been determined to 2.2 A resolution. The polypeptide folds in an unusual way to give four distinct domains enclosing a hole large enough to accommodate a double-stranded DNA. The active-site tyrosyl residue, which is involved in the transient breakage of a DNA strand and the formation of a covalent enzyme-DNA intermediate, is present at the interface of two domains. The structure suggests a plausible mechanism by which E. coli DNA topoisomerase I and other members of the same DNA topoisomerase subfamily could catalyse the passage of one DNA strand through a transient break in another strand.
Organizational Affiliation:
Department of Biochemistry, Molecular Biology and Cell Biology, Northwestern University, Evanston, Illinois 60208.