NMR structure and mutagenesis of the Fas (APO-1/CD95) death domain.
Huang, B., Eberstadt, M., Olejniczak, E.T., Meadows, R.P., Fesik, S.W.(1996) Nature 384: 638-641
- PubMed: 8967952
- DOI: https://doi.org/10.1038/384638a0
- Primary Citation of Related Structures:
1DDF - PubMed Abstract:
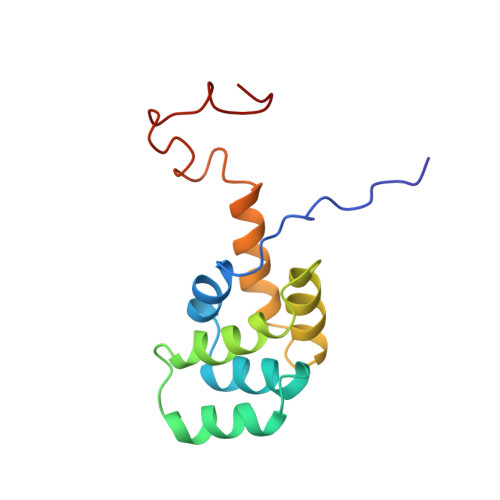
Programmed cell death (apoptosis) mediated by the cytokine receptor Fas is critical for the removal of autoreactive T cells, the mechanism of immune privilege, and for maintenance of immune-system homeostasis. Signalling of programmed cell death involves the self-association of a conserved cytoplasmic region of Fas called the death domain and interaction with another death-domain-containing protein, FADD (also known as MORT1). Although death domains are found in several proteins, their three-dimensional structure and the manner in which they interact is unknown. Here we describe the solution structure of the Fas death domain, as determined by NMR spectroscopy. The structure consists of six antiparallel, amphipathic alpha-helices arranged in a novel fold. From the structure and from site-directed mutagenesis, we have identified the region of the death domain involved in self-association and binding to the downstream signalling partner FADD.
Organizational Affiliation:
Pharmaceutical Discovery Division, Abbott Laboratories, Abbott Park, Illinois 60064, USA.