Sequential calcium binding to the regulatory domain of calcium vector protein reveals functional asymmetry and a novel mode of structural rearrangement.
Theret, I., Baladi, S., Cox, J.A., Sakamoto, H., Craescu, C.T.(2000) Biochemistry 39: 7920-7926
- PubMed: 10891072
- DOI: https://doi.org/10.1021/bi000360z
- Primary Citation of Related Structures:
1C7V, 1C7W - PubMed Abstract:
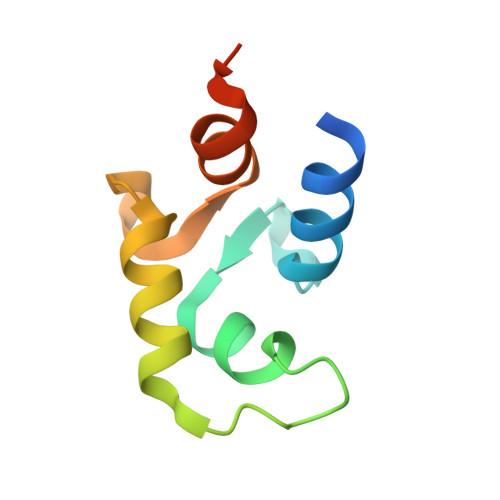
Calcium vector protein (CaVP) from amphioxus is a two-domain, calcium-binding protein (18.3 kDa) of the calmodulin superfamily. Only two of the four EF-hand motifs (sites III and IV) have a significant binding affinity for calcium ions. We determined the solution structure of the domain containing these active sites (C-CaVP: W81-S161), in the Ca(2+)-saturated state, using NMR spectroscopy and restrained molecular dynamics. The tertiary structure is similar to other Ca(2+)-binding domains containing a pair of EF-hand motifs. The apo state has spectroscopic and thermodynamic characteristics of a molten globule, with conserved secondary structure but highly fluctuating tertiary organization. Titration of C-CaVP with Ca(2+) revealed a stepwise ion binding, with a stable equilibrium intermediate in which only site III binds a calcium ion. Despite a highly fluctuating structure of the free site IV, the calcium-bound site III has a persistent structure, with similar secondary elements but different interhelix angle and hydrophobic packing relative to the fully calcium-saturated state.
Organizational Affiliation:
INSERM U350, Institut Curie-Recherche, Centre Universitaire, Bâtiments 110-112, 91405 Orsay, France.