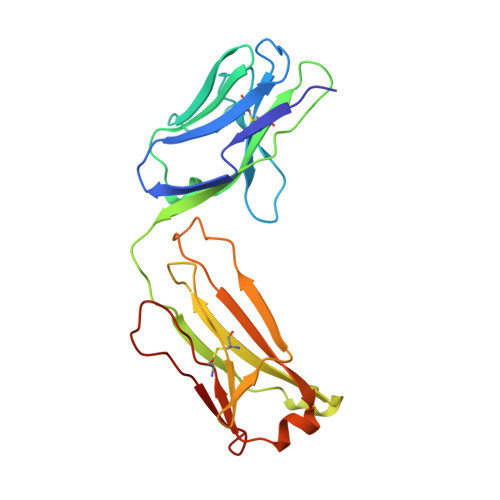
The crystal structure of the Fab fragment of a rat monoclonal antibody against the main immunogenic region of the human muscle acetylcholine receptor.
Kontou, M., Leonidas, D.D., Vatzaki, E.H., Tsantili, P., Mamalaki, A., Oikonomakos, N.G., Acharya, K.R., Tzartos, S.J.(2000) Eur J Biochem 267: 2389-2397
- PubMed: 10759865
- DOI: https://doi.org/10.1046/j.1432-1327.2000.01252.x
- Primary Citation of Related Structures:
1C5D - PubMed Abstract:
The crystal structure of the Fab fragment of a rat monoclonal antibody, number 192, with a very high affinity (Kd = 0.05 nM) for the main immunogenic region of the human muscle acetylcholine receptor (AChR), has been determined and refined to 2.4 A resolution by X-ray crystallographic methods. The overall structure is similar to a Fab (NC6.8) from a murine antibody, used as a search model in molecular replacement. Structural comparisons with known antibody structures showed that the conformations of the hypervariable regions H1, H2, L1, L2, L3 of Fab192 adopt the canonical structures 1, 1, 2, 1, and 1, respectively. The surface of the antigen-binding site is relatively planar, as expected for an antibody against a large protein antigen, with an accessible area of 2865 A2. Analysis of the electrostatic surface potential of the antigen-binding site shows that the bottom of the cleft formed in the center of the site appears to be negatively charged. The structure will be useful in the rational design of very high affinity humanized mutants of Fab192, appropriate for therapeutic approaches of the model autoimmune disease myasthenia gravis.
Organizational Affiliation:
Department of Biochemistry, Hellenic Pasteur Institute, Athens, Greece.