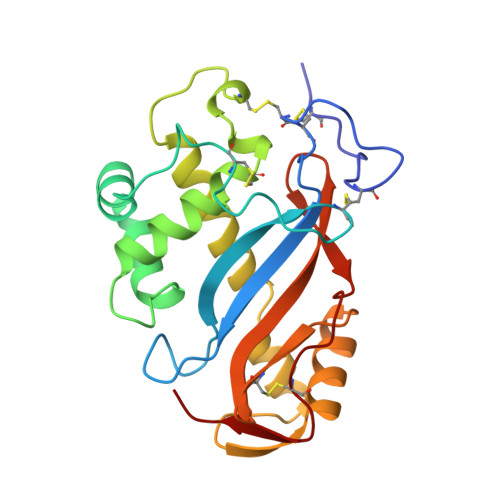
The crystal structure of ribonuclease Rh from Rhizopus niveus at 2.0 A resolution.
Kurihara, H., Nonaka, T., Mitsui, Y., Ohgi, K., Irie, M., Nakamura, K.T.(1996) J Mol Biol 255: 310-320
- PubMed: 8551522
- DOI: https://doi.org/10.1006/jmbi.1996.0025
- Primary Citation of Related Structures:
1BOL - PubMed Abstract:
The three-dimensional structure of ribonuclease Rh (RNase Rh), a new class of microbial ribonuclease from Rhizopus niveus, has been determined at 2.0 A resolution. The overall structure of RNase Rh is completely different from those of other previously studied RNases, such as RNase A from bovine pancreas and RNase T1 from Aspergillus oryzae. In the structure of RNase Rh, two histidine residues (His46 and His109) and one glutamic acid residue (Glu105), which were predicted to be critical to the activity from the chemical modification and mutagenesis experiments, are found to be located close together, constructing the active site. The indole ring of Trp49 plays an important role in preserving the active site structure by its stacking interactions with the imidazole ring of His 109, and by hydrogen bonding with the carboxyl group of Glu105. There exists a hydrophobic pocket around the active site, which contains the aromatic side-chain of Trp49 and Tyr57. The results of mutagenesis studies suggest that this pocket is the base binding site of the substrate.
Organizational Affiliation:
Department of BioEngineering, Nagaoka University of Technology, Japan.