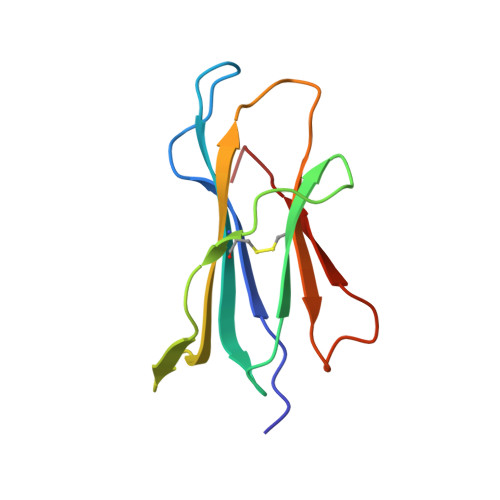
Three-dimensional structure of beta 2-microglobulin.
Becker, J.W., Reeke Jr., G.N.(1985) Proc Natl Acad Sci U S A 82: 4225-4229
- PubMed: 3889925
- DOI: https://doi.org/10.1073/pnas.82.12.4225
- Primary Citation of Related Structures:
1BMG - PubMed Abstract:
The three-dimensional structure of beta 2-microglobulin, the light chain of the major histocompatibility complex class I antigens, has been determined by x-ray crystallography. An electron density map of the bovine protein was calculated at a nominal resolution of 2.9 A by using the methods of multiple isomorphous replacement and electron density modification refinement. The molecule is approximately 45 X 25 X 20 A in size. Almost half of the amino acid residues participate in two large beta structures, one of four strands and the other of three, linked by a central disulfide bond. The molecule thus strongly resembles Ig constant domains in polypeptide chain folding and overall tertiary structure. Amino acid residues that are the same in the sequences of beta 2-microglobulin and Ig constant domains are predominantly in the interior of the molecule, whereas residues conserved among beta 2-microglobulins from different species are both in the interior and on the molecular surface. In the crystals studied, the molecule is clearly monomeric, consistent with the observation that beta 2-microglobulin, unlike Ig constant domains, apparently does not form dimers in vivo but associates with the heavy chains of major histocompatibility complex antigens. Our results demonstrate that, at the level of detailed three-dimensional structure, the light chain of the major histocompatibility class I antigens belongs to a superfamily of structures related to the Ig constant domains.