Crystallographic Analysis of Transition State Mimics Bound to Penicillopepsin: Difluorostatine-and Difluorostatone-Containing Peptides
James, M.N.G., Sielecki, A.R., Moult, J.(1992) Biochemistry 31: 3872
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
(1992) Biochemistry 31: 3872
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| PROTEIN (PENICILLOPEPSIN) | A [auth E] | 323 | Penicillium janthinellum | Mutation(s): 0 EC: 3.4.23.20 | 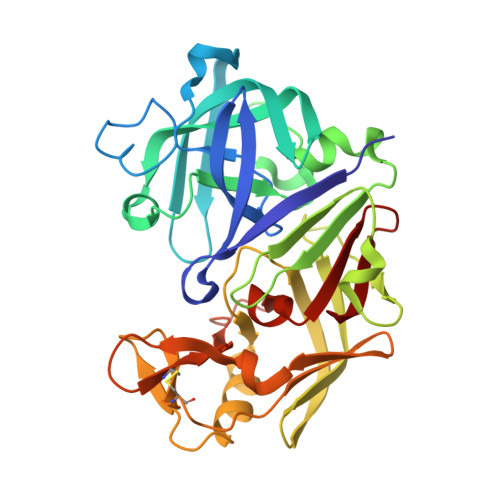 |
UniProt | |||||
Find proteins for P00798 (Penicillium janthinellum) Explore P00798 Go to UniProtKB: P00798 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P00798 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Find similar proteins by: Sequence | 3D Structure
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| PEPSTATIN ANALOGUE ISOVALERYL-VAL-VAL-STA-O-ET | B [auth I] | 4 | N/A | Mutation(s): 0 | 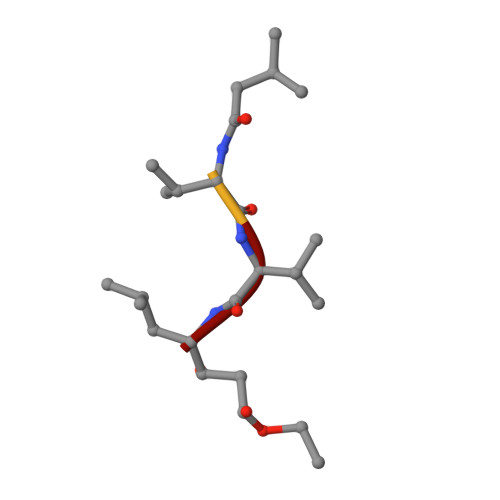 |
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| MAN Query on MAN | C [auth I] | alpha-D-mannopyranose C6 H12 O6 WQZGKKKJIJFFOK-PQMKYFCFSA-N |  | ||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| ID | Chains | Name | Type/Class | 2D Diagram | 3D Interactions |
| PRD_001238 Query on PRD_001238 | B [auth I] | PEPSTATIN ANALOGUE ISOVALERYL-VAL-VAL-STA-O-ET | Oligopeptide / Enzyme inhibitor |  | |
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 97.24 | α = 90 |
| b = 46.5 | β = 115.34 |
| c = 65.65 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PROLSQ | refinement |